Theory of aqueous solubility prediction
Table of contents
This page summarizes the theoretical background behind Chemaxon's Solubility (logS) Predictor. To find more information about the predictor, see the following page.
Introduction
Aqueous solubility is one of the most important physico-chemical properties in modern drug discovery. It has impact on ADME-related properties like drug uptake, distribution and even oral bioavailability. Solubility can also be a relevant descriptor for property-based computational screening methods in drug discovery process. Hence there is a significant interest in fast, reliable methods for predicting solubility in water for promising drug candidates.
Chemaxon's Solubility Predictor is able to calculate two types of solubility: intrinsic and pH-dependent solubility.
LogS is a common unit for measuring solubility. It is the 10-based logarithm of the solubility measured in mol/l unit, so logS = log (solubility measured in mol/l).
The Solubility Predictor predicts solubility values at 25 °C.
The predictor can provide quantitative results, calculating solubility in logS, mg/mL or mol/L units. The predictive accuracy of the plugin is considered to be 1 logS unit. In case a qualitative estimation of the solubility is needed, the plugin can return a solubility category.
Intrinsic solubility
The intrinsic solubility (usually denoted as logS0) of an ionizable compound is the solubility that can be measured after an equilibrium of solvation between the dissolved and the solid state is reached at a pH where the compound is fully neutral.
Example
The intrinsic solubility of phenol can be measured at pH 6.0. Phenol is a weak acid with a pKa value of 10.02, which means that at pH 6.0 the molecule is dominantly present in its neutral form. So at pH 6.0 equilibrium can be measured only between the solid and the dissolved neutral form.
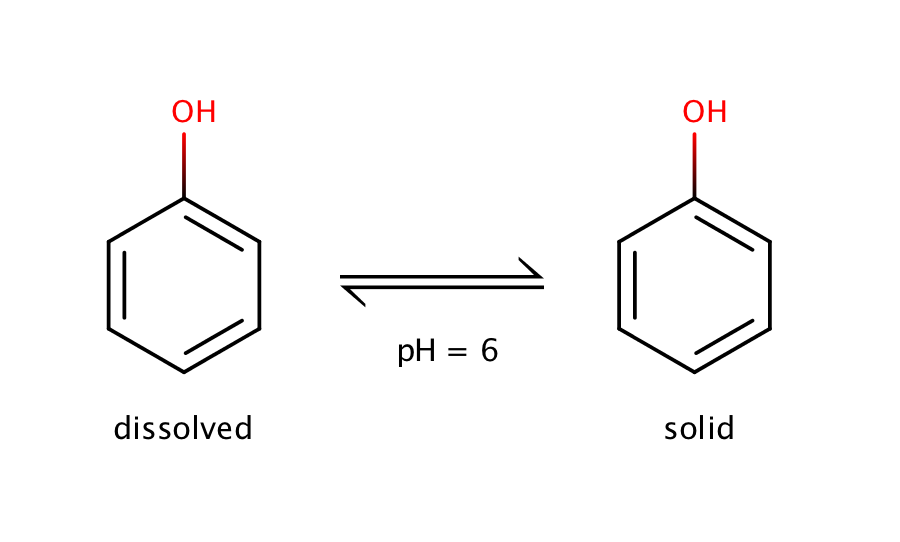
Fig. 1. Solvation equilibrium of phenol at pH 6.0
Our predictor uses a fragment-based method that identifies different structural fragments in the molecule and assigns an intrinsic solubility contribution to them. The contributions then are summed up to determine the intrinsic solubility value. The implementation of the fragment-based method is based on the article of Hou et al.
The figure below shows a molecule split up into fragments that are used in the intrinsic solubility prediction.
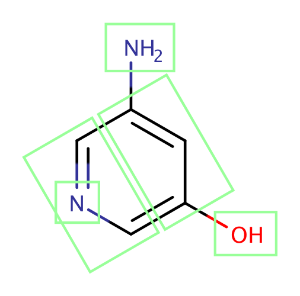
Fig. 2. A molecule split into fragments to predict its intrinsic solubility
pH-dependent solubility
The pH of a solution affects the ionization of the dissolved compound, shifting its solvation equilibrium. With increasing ionization solubility increases compared to the intrinsic solubility.
Example
The solubility of aniline in an acidic environment will be greater than its intrinsic solubility as the protonation of the compound shifts the equilibrium between the pure liquid aniline and its dissolved form to the right side (to the dissolved form).
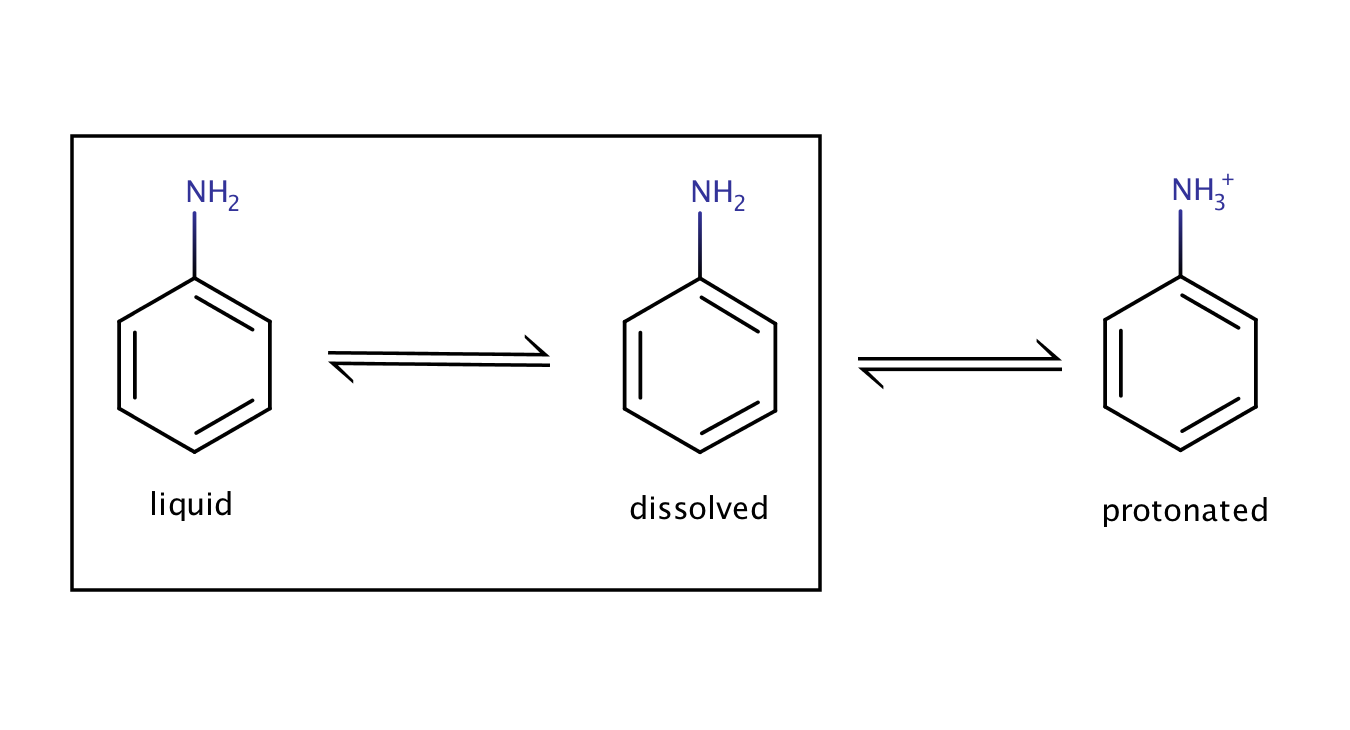
Fig. 3. Solvation equilibrium of aniline in an acidic environment
The pH-dependent solubility (usually denoted as logSpH) can be derived from the Henderson-Hasselbalch equation and the above definition of intrinsic solubility.
In case of a weak acid the formula is the following:
logSpH = logS0 + log(1 + 10(pH-pKa))
Based on the derivations for mono-/di-protic acids/bases and ampholytes, the formula above can be transformed into the following general one:
logSpH = logS0 + log(1 + α), where α = ∑iαcAi/∑jαnAj
In this formula αcAi is the distribution % of the i-th charged microspecies at the given pH, while αnAj is the distribution % of the j-th neutral microspecies at the given pH.
Example
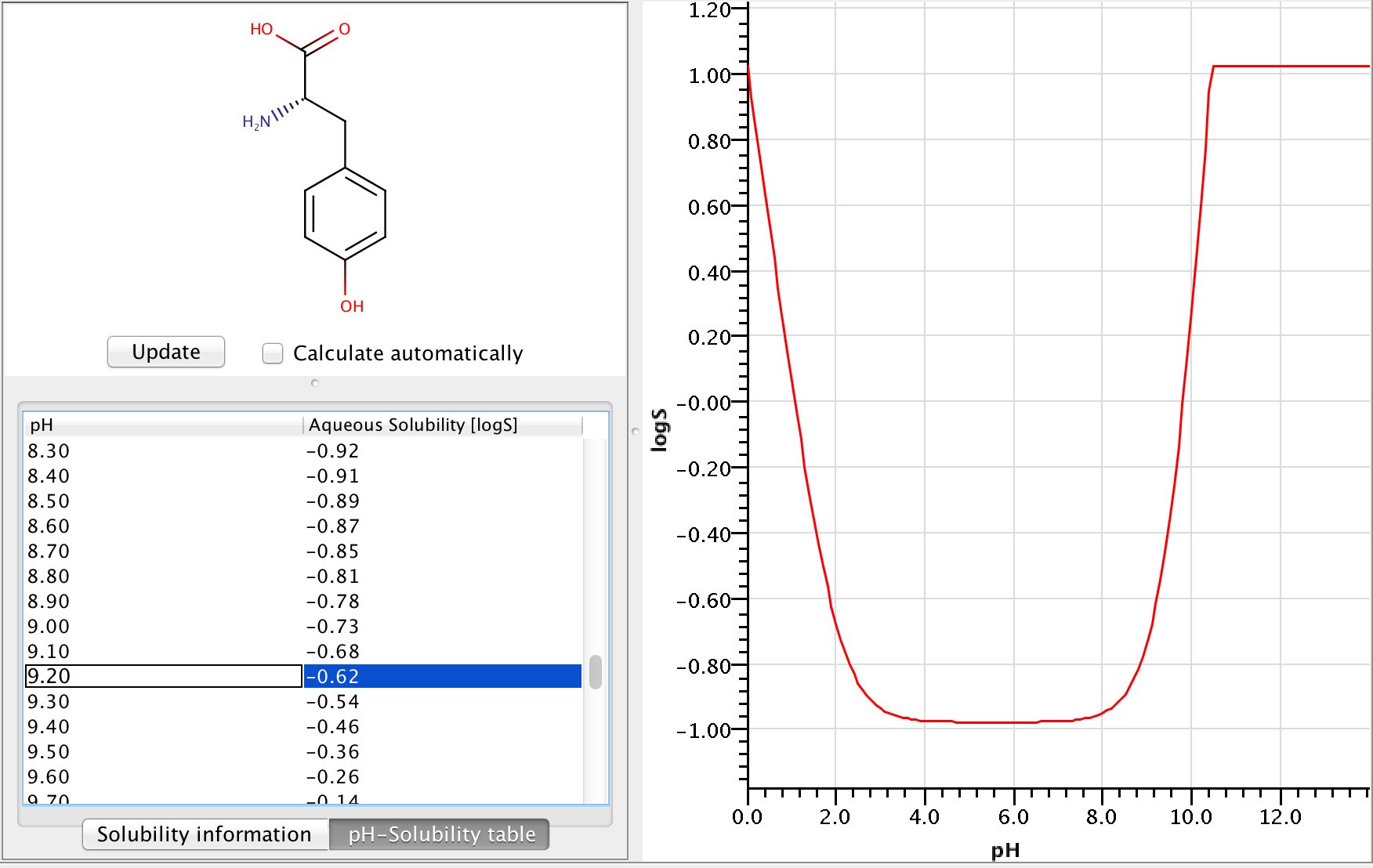
Let's calculate the pH-dependent solubility of L-tyrosine at pH=9.2.
Zwitterionic molecules are the least soluble around their isoelectric point. The predicted isoelectric point of L-tyrosine is 5.5, which means that we can expect better solubility at pH 9.2 than at pH 5.5.
To get the pH-dependent solubility we first need the instrisic solubility of L-tyrosine. The logS Predictor predicts -0.98 logS as intrinsic solubility.
To take ionization into account we have to calculate the microspecies distribution of L-tyrosine at pH=9.2. To do this we will use the pKa calculator, which can calculate the microspecies distribution based on the predicted pKa values. The following image shows the calculated distributions, with the highlighted row showing the distributions at pH 9.2.
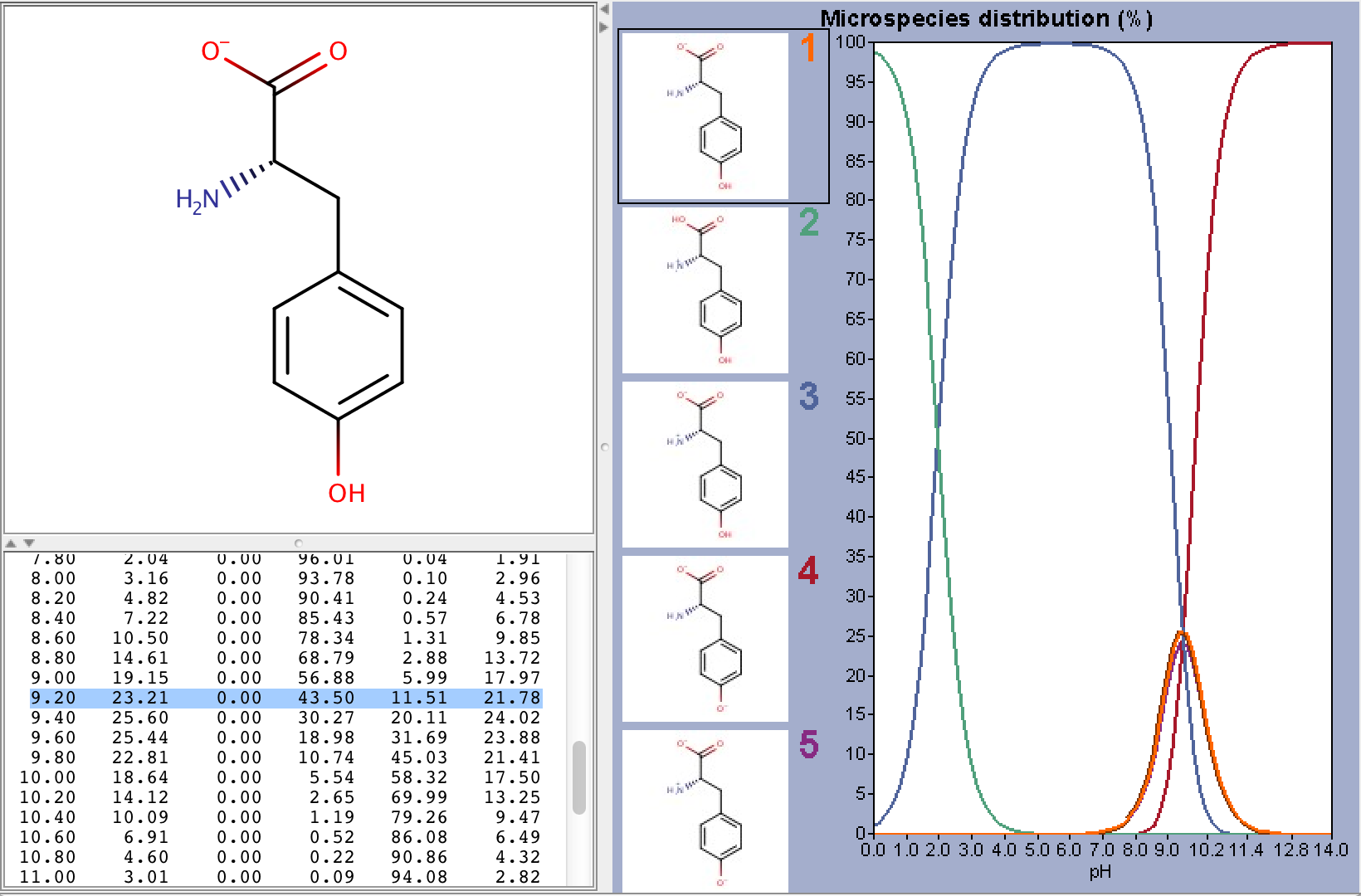
From the image above we can read that the distribution %s of the charged microspecies are (with their charges shown):
23.21 (-1), 0.0 (+1), 11.51 (-1, 1), 21.78 (-1, -1, +1)
The distribution % of the neutral microspecies:
43.50 (zwitterionic species)
Using these we can easily calculate the log(1 + α) correction:
log(1 + α) = log(1 + (23.21 + 0.0 + 11.51 + 21.78)/43.5) = 0.362
Adding the correction to the intrinsic solubility, we get that the pH-dependent solubility at pH 9.2 is -0.618 logS.
The following image shows the whole pH-logS curve of tyrosine with the calculated solubility at pH 9.2.
Cut-off of the pH-dependent solubility curve
To put practical limits to the pH-dependent solubility curve and describe the fact that the solution reaches a certain saturation, a cut-off is applied to better match the real (experimental) pH-dependent solubility curve.
In our logS Predictor we apply the following cut-off (the following solubility values are all expressed in logS unit):
-
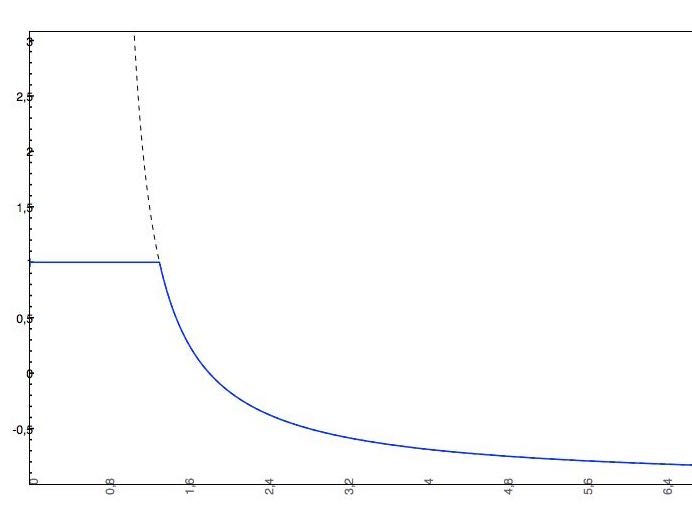
if the predicted logS0 > -2, the applied cut-off will be +2, which means that the pH-dependent logS curve will be "cut off" at logS0 + 2. This means that the pH-dependent logS values won't increase above logS0 + 2.
-
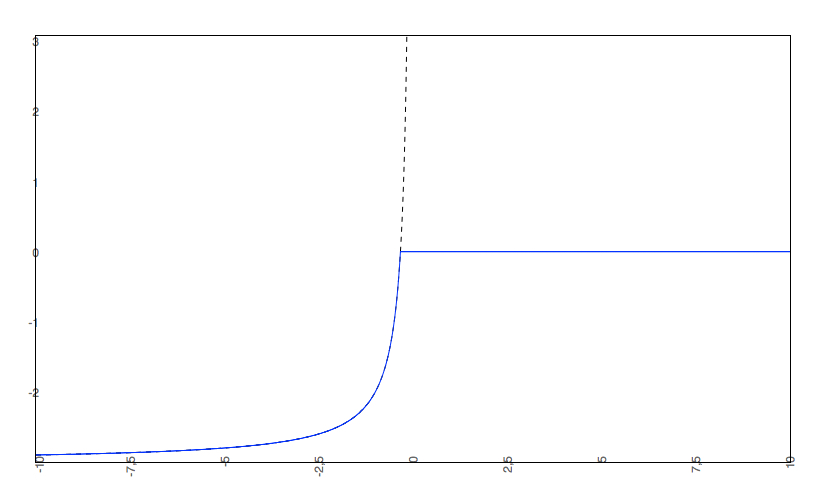
if the predicted logS0 < -2, the predicted pH-dependent solubility curve will not be allowed to rise above 0, so the cut-off will be at 0.
Cut-off examples
- The predicted intrinsic solubility (to which value the pH-dependent logS curve converges) is -1.0. Therefore the pH-dependent curve is cut off at +1.0.
- The predicted intrinsic solubility (to which the pH-dependent logS curve converges) is -3.0. Therefore the pH-dependent curve is cut off at 0.
Examples of predicted pH-dependent logS curves
In this section you will find some examples of predicted vs. experimental pH-dependent logS curves for specific drug molecules. The experimental values and the detailed analysis of the experimental logS curves can be found in the referenced literature.
Example 1
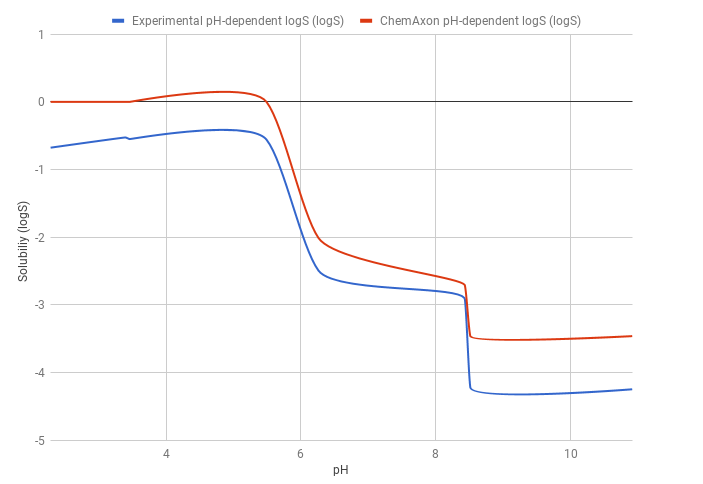
The predicted pH-dependent logS curve of the HCl salt of ticlopidine shows a good correlation with the experimental curve, meaning that the HH-equation quite accurately describes the pH-dependence of the solubility. This result also comes from the fact that ticlopidine is a monoprotic base, so the HH-equation works well.
The predicted logS0 is -3.47, while the experimental logS0 is -4.25. The absolute difference between the experimental and predicted intrinsic solubility is 0.78 logS unit.
Example 2
The predicted pH-dependent logS curve of the hydrogen fumarate salt of the diprotic base quetiapine shows good correlation with the experimental curve. Its predicted logS0 is-4.27, while the experimental logS0 -2.84. The absolute difference between the experimental and the predicted instrinsic solubility is 1.42.
Example 3
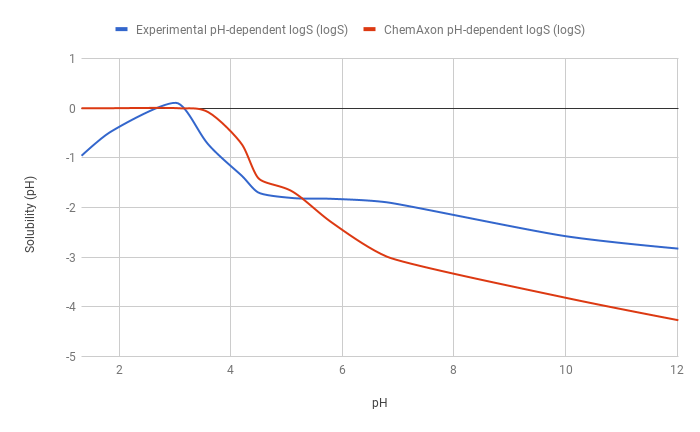
The predicted pH-dependent logS curve of the hydrogen fumarate salt of the ampholytic desvenlafaxine shows difference to the experimental curve below pH 6, which is due to salt formation. The predicted intrinsic logS is -2.16, while the experimental is -3.25. The absolute difference between these two values is 1.09 logS unit.
Results of testing the model
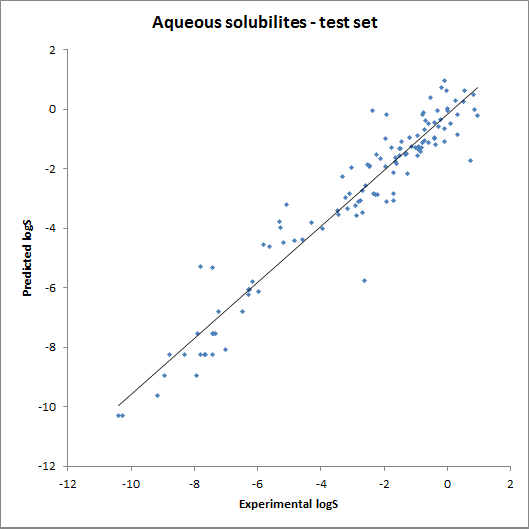
The accuracy of the model was tested using fragment contributions calculated from the training set with a linear regression model. The obtained contributions were then used for calculating solubility for the training and the test set. The results are summarised on the following two charts:

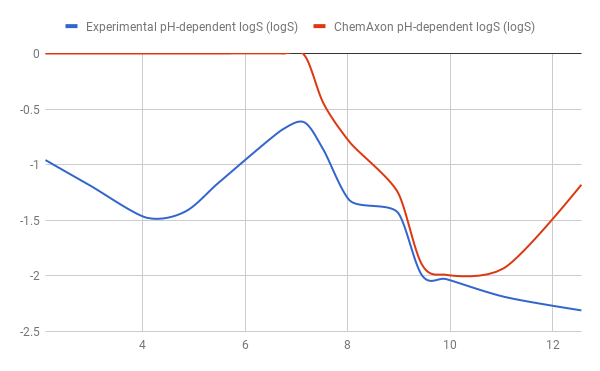
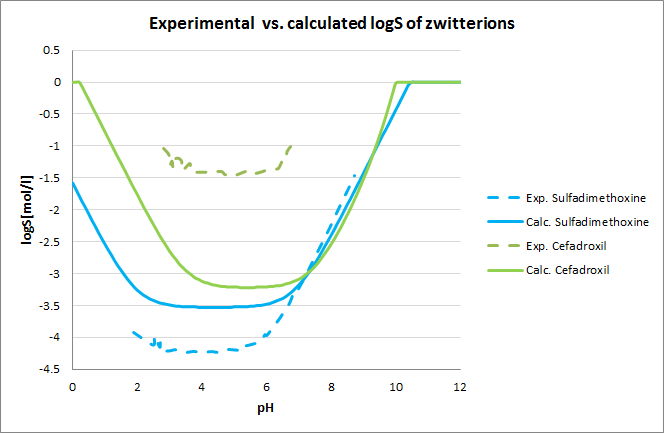
Tests for pH-logS profile were also run. The two plots below show calculated and experimental pH-logS profiles for different acidic, basic and zwitter-ionic compounds:
The Solubility Predictor will be developed further in the future. Among our future goals we have extending the prediction with a descriptor-based method and adding training features.
References
1. Hou, T. J.; Xia, K.; Zhang, W.; Xu, X. J. ADME Evaluation in Drug Discovery. 4. Prediction of Aqueous Solubility Based on Atom Contribuition Approach. J. Chem. Inf. Comput. Sci. 2004 , 44 , 266-275
2. Völgyi, G.; Baka, E. et al. Study of pH-dependent solubility of organic basis. Revisit of the Henderson-Hasselbalch relationship, Analytica Chimica Acta, 2010 , 673, 40-46
- Avdeef, A. et al. Equilibrium solubility measurement of ionizable drugs - consensus recommendations for improving data quality, ADMET & DMPK 4(2), 2016 , 117-178
4. Shoghi, E.; Fuguet, E.; Bosch, E.; Rafols, C. Solubility-pH profiles of some acidic, basic and amphoteric drugs, European Journal of Pharmaceutical Sciences 2013 , 48, 291-300