JChem Base and Cartridge Performance Information
-
Scalability
-
Hardware Requirements
-
Benchmarks
-
Tuning
Scalability
How scalable is JChem Cartridge / JChem Base?
What is the maximum number of structures for JChem Base / JChem Cartridge?
In a single table the maximum number of rows is 231-1 (2,147,483,647).
We may increase this in the future if needed.
There is no practical limit for the number of tables.
Hardware requirements
How can I estimate hardware requirements for JChem Base / JChem server for JChem Cartridge?
-
Memory:
JChem may use significant amount of memory for building a structure cache in memory.
Please see this topic on how to estimate the size of the structure cache.
The total Java memory consumption (heap size) consists of:
-
The size of the structure cache
-
Processing memory for JChem Base: we recommend to multiply the estimated cache size by a factor of 0.75 (up to 8 processor cores). In case of more than 8 cores you may have higher memory need since it scales up with the number of the threads started.
-
Processing memory for your Java application (built on JChem Base)
Cache and Java memory usage measured for different table size on a 8 core 64-bit Linux (version 3.2.0-48-generic) environment:
Target count Cache size measured (MB) Cache size estimated (MB) Java memory allocation (MB) 986,581 148.85 152.45 250 5,032,252 701 720 1,250 10,002,807 1,610 1,637 3,000 19,987,542 3,174 3,206 6,500 38,221,498 6,002 6,144 12,000
-
The Java memory usage could be further improved (compared to the table values) with fine-tuning the JVM parameters.
To determine the total amount of RAM requirement you should add:
-
Java memory consumption (detailed above)
-
Other programs running on that computer
-
Memory requirement for the operating system
64 bit systems: A single Java process cannot allocate more than around 2 GB on 32 bit systems. If your Java memory needs will exceed this limit, a 64 bit system is recommended (including hardware, operating system and Java).
-
Disk space:
Apart from the number of structures it also depends on the format of the input file, and type of RDBMS engine used by JChem Base.
A benchmark result with 1 million structures (the NCI dataset was multiplied for the test), the RDBMS was Oracle:
| Format | Size of JChem table | Original file size |
|---|---|---|
| SDF | 1.2 GB | 3.7 GB |
| SMILES | 270 MB | 50 MB |
Note: JChem compresses SDF in the cd_structure field by default (in the case of the NCI dataset to roughly about 3 times smaller). This can be disabled (e.g. if they need to be displayed directly by non-Chemaxon tools), but the storage size increases in this case.
-
Processing power:
Comparing the performance of different hardware architectures is a complex topic and not the subject of this FAQ.
Some quick facts:
-
JChem automatically uses all processors during a search
-
The search speed scales well with the number of processors
-
The search time is directly proportional to the size of the table (assuming there is no constraint on the number of hits - see below)
-
The following benchmarks can be used as a starting point
For the same database the search time greatly depends on the type of query structure. For a very general query (e.g. benzene) there will be a lot of hits, meaning longer execution time, while more specific queries run very fast on the same large database.
The search time consists of
-
Cache load time : the cache is built up during the very first search
-
Screening time : this is a quick pre-filtering using fingerprints
-
Graph search: slower, but only performed for the screened compounds
The following can be stated:
-
The screening time is directly proportional to the size of the database table.
-
The graph search time is directly proportional to the number of screened compounds.
-
The number of hits is roughly proportional to the number of of screened compounds.
-
The graph search time is roughly proportional to the number of hits.
Tip: it is rarely useful to return a huge number of hits (especially for human consumption).
If the number of hits is limited, only the rapid screening time will increase with table size, which means the total search time will remain almost constant regardless of the table size.
How to estimate the memory need for the Structure Cache?
Typically 1 million drug-like structures consume around 160 MB memory in the structure cache of JChem.
Note: although JChem can drop the least recently used table from the structure cache if low on memory, it is recommended that all structure tables should fit in the cache (as cache loading can take a considerable amount of time). When estimating the memory need simply sum the number of rows in the tables.
The following table shows typical memory needs for standard structures:
| Test specifications | |
|---|---|
| Number of molecules: | 10,000,000 |
| Fingerprint size: | 16*32=512 bits |
| Average SMILES length per molecule: | 60.4 |
| Memory consumption: | 1,604.1 MB |
| Caching time: | 900 seconds |
Memory need increases with the number of molecules, the size of fingerprints, and the average SMILES string size. The following approximation can be used when number_of_molecules > 25000:
memory_need[bytes] =
number_of_molecules * (0.5 * average_smiles_length[characters] + fingerprint_size[bits] / 8 + 74)The structure table fingerprint statistics generation function can be used to report the average smiles length and fingerprint size of a JChem table or JChem index. See more information in the JChem Manager command line usage (s command), at the Cartridge index statistics function and the Statistics tab at Instant JChem Schema editor.
Benchmarks
How fast is importing in JChem Base?
The following table shows the duration of import in some cases. The configuration was the same as in the cartridge benchmark.
| Number of structures | Elapsed Time (ms) | |
|---|---|---|
| Duplicates allowed | Duplicates not allowed | |
| 10000 | 40406.0 | 44923.0 |
| 100000 | 405781.0 | 430840.0 |
| 200000 | 824870.0 | 883681.0 |
Notes:
-
If duplicates are allowed, time increases linearly with the number of molecules imported.
-
If duplicates are not allowed, JChem performs a search for every molecule to check if it is already in the structure table or not.
-
The table into which the structures were imported always contained an initial 10 structures and were stats-collected
How fast is substructure searching in JChem Base?
The following tests demonstrate the speed of substructure search in JChem.
The test configuration was the same as in the cartridge benchmark, and the same query structures were used.
-
A chemical table containing 38,165,924 molecules was used;
-
Fingerprints and SMILES were cached by JChem;
-
JChem version: 6.3.0;
-
Java version: Oracle Corporation 1.7.0_17;
-
OS: amd64 Linux 2.6.18-164.0.0.0.1.el5xen.
Query Structure Search Options Number of Hits Screened Count Search Time (ms) 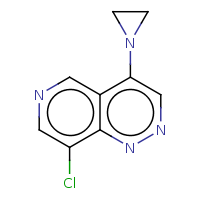
t:s 0 0 3116.0 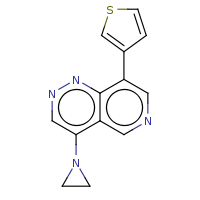
t:s 0 0 3132.0 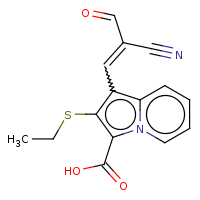
t:s 3 3 3506.0 
t:s 77 115 3157.0 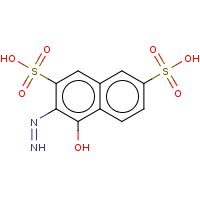
t:s 93 193 2780.0 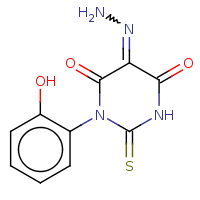
t:s 117 117 2775.0 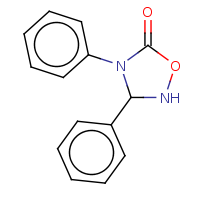
t:s 137 137 2787.0 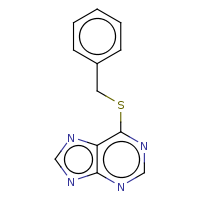
t:s 696 720 3153.0 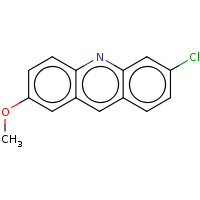
t:s 1127 1337 3570.0 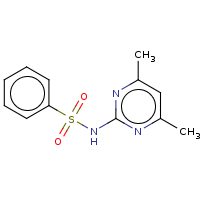
t:s 3841 3973 3155.0 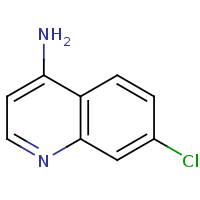
t:s 10047 10421 3647.0 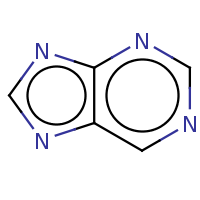
t:s 233472 233852 5620.0 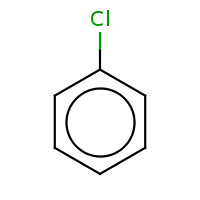
t:s 5937200 5990786 76444.0 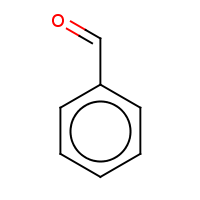
t:s 8339436 9189891 129767.0
The column names have the following meaning:
-
Query Structure : The query tested.
-
Search Options : The search options applied.
-
Number of Hits : The number of structures returned by the query.
-
Screened Count : The number of structure left over from the fingerprint screening as possible candidates meeting the search criteria.
How fast is inserting in JChem Cartridge?
The following table shows the duration of inserting in some cases. The configuration was exactly the same as in the cartridge benchmark.
| Number of structures | Elapsed Time (ms) | |
|---|---|---|
| Duplicates allowed | Duplicates not allowed | |
| 10000 | 169040.0 | 191999.0 |
| 100000 | 1684728.0 | 3101339.0 |
| 200000 | 3555535.0 | 9296357.0 |
How fast is indexing in JChem Cartridge?
The following table shows the duration of indexing in some cases. The configuration was exactly the same as in the cartridge benchmark.
| Number of structures | Elapsed Time (ms) | |
|---|---|---|
| Duplicates allowed | Duplicates not allowed | |
| 10000 | 12602.0 | 14881.0 |
| 100000 | 72729.0 | 782445.0 |
| 200000 | 145654.0 | 3500274.0 |
How fast is searching in JChem Cartridge?
The following table shows the duration of search in JChem Cartridge using the following configuration:
-
JChem version: 6.3.0;
-
Oracle version: Oracle Database 11g Enterprise Edition Release 11.2.0.3.0 - 64bit;
-
Hardware: Amazon EC2, m2.4xlarge instance, EBS storage;
-
Target molecule set: PubChem;
-
Number of structures: 38,165,924.
Session Date : 2013-10-05
Query Structure Search Options Number of Hits Screened Count Search Time (ms) 
t:s earlyResults:2000 0 0 2803.0 
t:s earlyResults:2000 0 0 3187.0 
t:s earlyResults:2000 3 3 3512.0 
t:s earlyResults:2000 77 115 2835.0 
t:s earlyResults:2000 93 193 3181.0 
t:s earlyResults:2000 117 117 2802.0 
t:s earlyResults:2000 137 137 3151.0 
t:s earlyResults:2000 696 720 3598.0 
t:s earlyResults:2000 1127 1337 3210.0 
t:s earlyResults:2000 3841 3973 3601.0 
t:s earlyResults:2000 10047 10421 3337.0 
t:s earlyResults:2000 233472 233852 6258.0 
t:s earlyResults:2000 5937200 5990786 86830.0 
t:s earlyResults:2000 8339436 9189891 143022.0 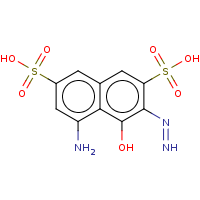
t:t simThreshold:0.9 0 0 4209.0 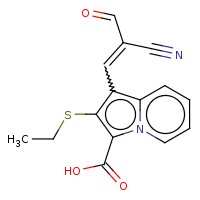
t:t simThreshold:0.9 0 0 4969.0 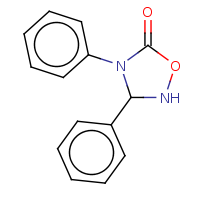
t:t simThreshold:0.9 0 0 2272.0 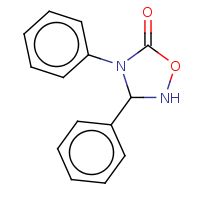
sep=! t:s!ctFilter:(PSA() <= 200) && (rotatableBondCount() <= 10) && (mass() <= 500) && (aromaticRingCount() <= 4) 130 137 3011.0 
sep=! t:s!ctFilter:(mass() <= 500) && (logP() <= 5) && (donorCount() <= 5) && (acceptorCount() <= 10) 15 137 2962.0
The column names have the following meaning:
-
Query Structure : The query tested.
-
Search Options : The name of the operator tested.
-
Number of Hits : The number of structures returned by the query.
-
Screened Count : The number of structure left over from the fingerprint screening as possible candidates meeting the search criteria.
-
Search Time : The total time spent executing the SQL statement.
Tuning
What can/should I do to make JChem Cartride searches faster?
You can find a few performance tuning hints here.