Markush Editor User's Guide
Contents
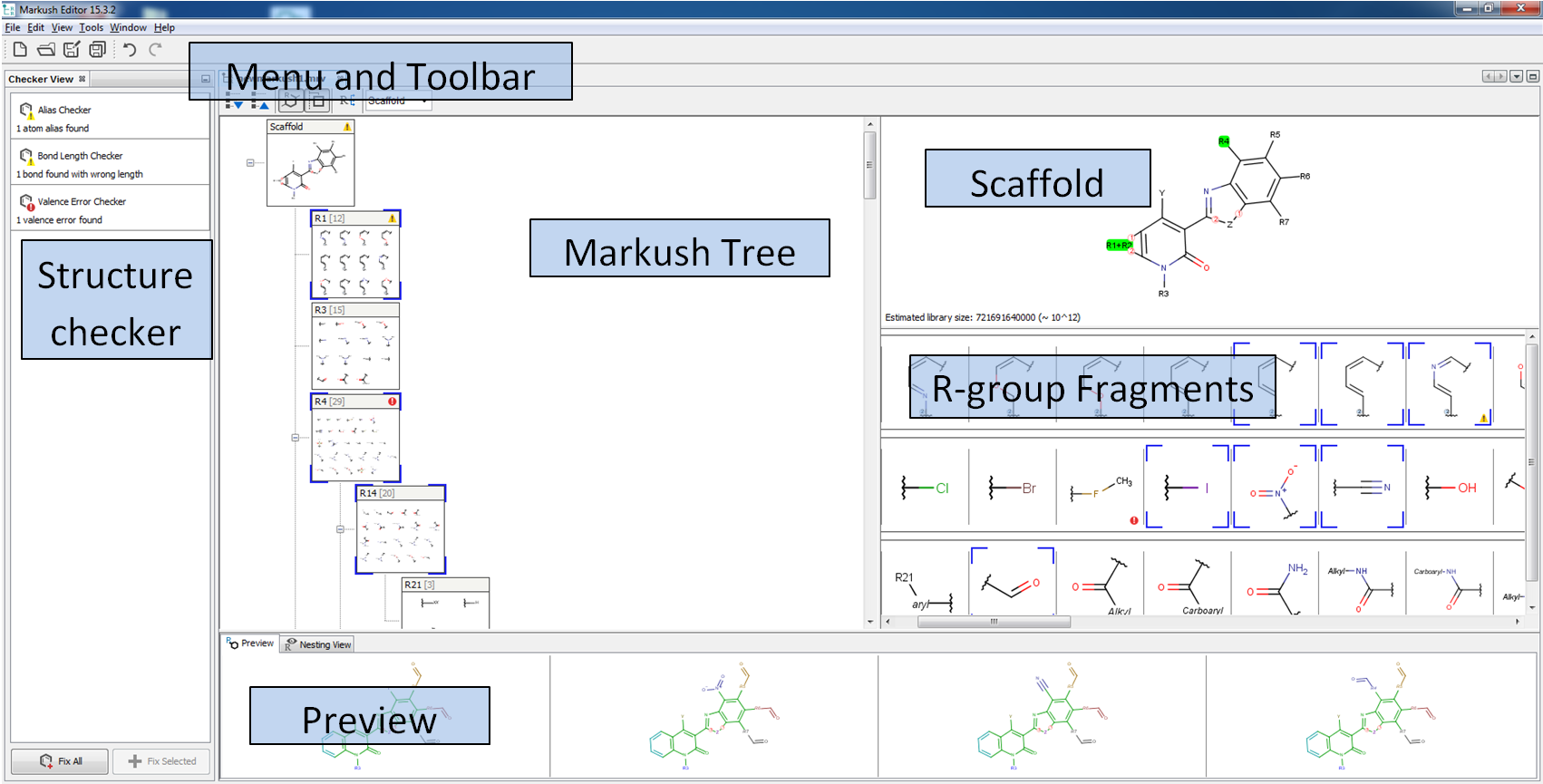
Markush Editor Application
![]() Markush Editor is a desktop application of ChemAxon that serves for viewing, creating and editing complex Markush structures. Markush Editor uses a graphical representation of the hierarchical nature of the R-groups in Markush structures. The structures (fragments) of an opened Markush file will be identified, classified and organized as scaffold and attached R-groups to help you follow each nested structure. Markush Editor is a standalone desktop application. For running this application, you need to download and run Markush Editor installer. Without a valid Markush Editor license, the application can only be used in viewer mode. Short video tutorials demonstrating the main functionalities of the application are available here.
Markush Editor is a desktop application of ChemAxon that serves for viewing, creating and editing complex Markush structures. Markush Editor uses a graphical representation of the hierarchical nature of the R-groups in Markush structures. The structures (fragments) of an opened Markush file will be identified, classified and organized as scaffold and attached R-groups to help you follow each nested structure. Markush Editor is a standalone desktop application. For running this application, you need to download and run Markush Editor installer. Without a valid Markush Editor license, the application can only be used in viewer mode. Short video tutorials demonstrating the main functionalities of the application are available here.
Main Menu and Toolbar
The main menu contains File , View , Tools , Window and Help elements.

-
File
-
New,

-
Open (Ctrl+O),

-
Compose Markush Structure...,

-
Save,

-
Save As...,

-
Save All,

-
Export to Text...,
-
Exit.
-
-
View
-
Show Only Editor (Ctrl+Shift+Enter)
-
Full Screen (Alt+Shift+Enter)
-
-
Tools
-
Plugins
-
Options...

-
-
Windows
-
Checker View
-
Reset Windows
-
Close Window
-
Close All Documents
-
Close Other Documents
-
Documents...
-
-
Help
-
Help (F1)

-
Licenses...

-
Check for Updates...
-
About ChemCurator...
-

-
"Collapse molecule tree"
 Closes all nodes of the R-group tree. The default setting is off .
Closes all nodes of the R-group tree. The default setting is off . -
"Expand molecule tree"
 Expands all nodes of the R-group tree. The default setting is off .
Expands all nodes of the R-group tree. The default setting is off . -
"Always display scaffold in Fragments"
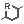 Displays the relevant scaffold of the selected R-group. The default setting is on .
Displays the relevant scaffold of the selected R-group. The default setting is on . -
"Toggle molecule display in tree"
 Expands the boxes of the tree to show the structure of the molecule. The default setting is on .
Expands the boxes of the tree to show the structure of the molecule. The default setting is on . -
R-group search box: allows quick navigation the the desired R-group of the Markush structure.
-
"Enumerate Markush structure"
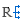 Open the Markush enumeration panel.
Open the Markush enumeration panel.
Panels and views
Markush Editor contains five main components by default. All of the components can be reorganized and resized.
-
Editor panel contains Markush tree, Scaffold and R-group fragments view;
-
Checker checker displays the error and warnings related your Markush structure;
-
Preview displays enumerated examples;
-
Nesting view displays the embedding of the selected R-group(s);
-
Validation view can display the validation status (matching or non-matching) of individual compounds against your Markush structure.
Editor panel
The Markush structures of the opened file are organized and arranged in the Editor panel. In the tree view, the scaffold is presented on the first (parent) level of the tree, and the associated R-groups are classified as its children. The scaffold(s) and attached groups are denoted by named boxes. The parent level boxes are named as "Scaffold". Nested nodes are labeled according to the represented R-group definitions (e.g.: R1, R2,...Rn or the alias of the R-group if it has one), followed by the number of members. To view any scaffold or R-group definition, select the relevant node. The structure(s) of the chosen element will be presented in the fragment view. The selected items are highlighted in the tree view.
Select a node and see the relevant structure(s) in the View panel
You can select more than one structure from the tree using Shift+Left-Click or Ctrl+Left-Click to display them in the View panel at the same time.
Note: Shift/Ctrl+Arrow keys work for selection as well.
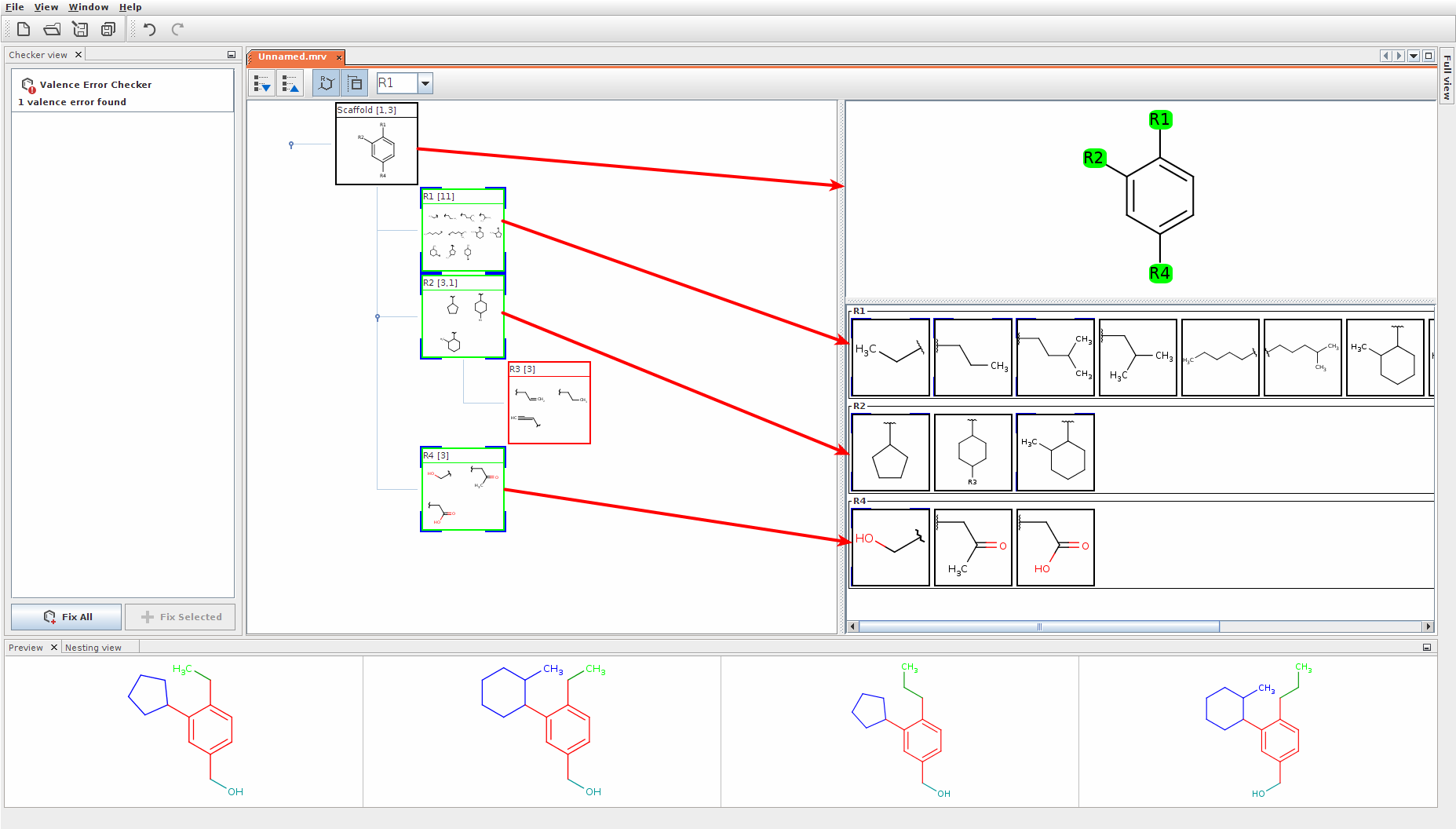
Select more nodes and see structures in the View panel
Expand the nodes of the Tree panel to show the small structures of each node.
Preview panel
Preview tab displays enumerated examples (max. 4) of the selected R-group(s).
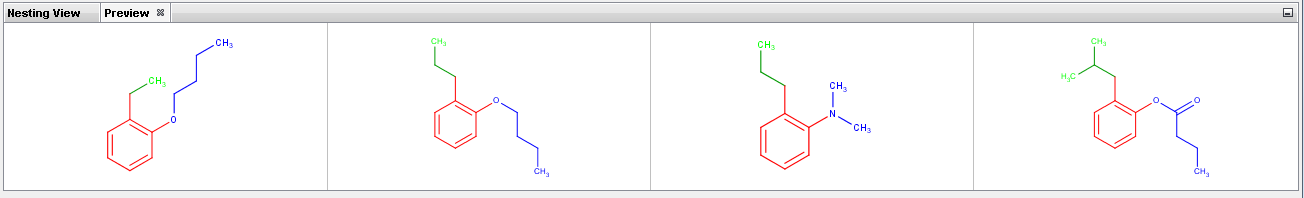
See enumerated examples in Preview panel
Nesting view panel
Nesting view tab displays the step-by-step route of embedding of the selected R-group starting from the scaffold.
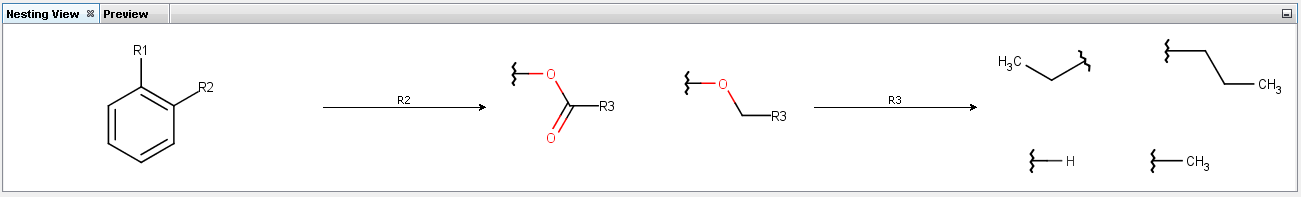
Follow the embedding of the selected R-group in Nesting panel
Validation panel
Validation panel can be used to check whether individual compounds match your Markush structure or not. Matching structures are highlighted by a green background, while non-matching structures are highlighted in red.
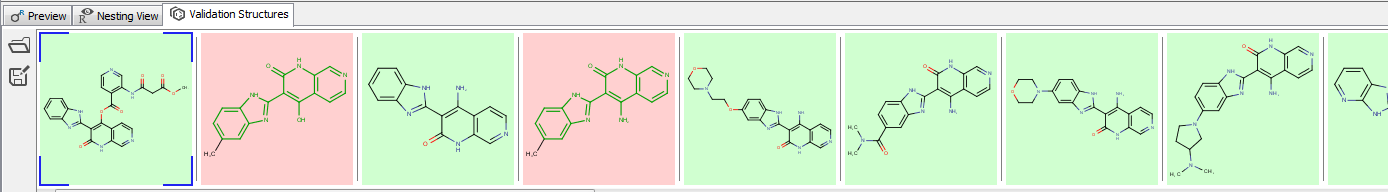
Display of matching and non- matching structures in Validation panel
If any of the validation structures are selected, the matching and non-matching R-groups and fragments are also highlighted in Markush structure (in the scaffold view, the tree view and the fragment view).
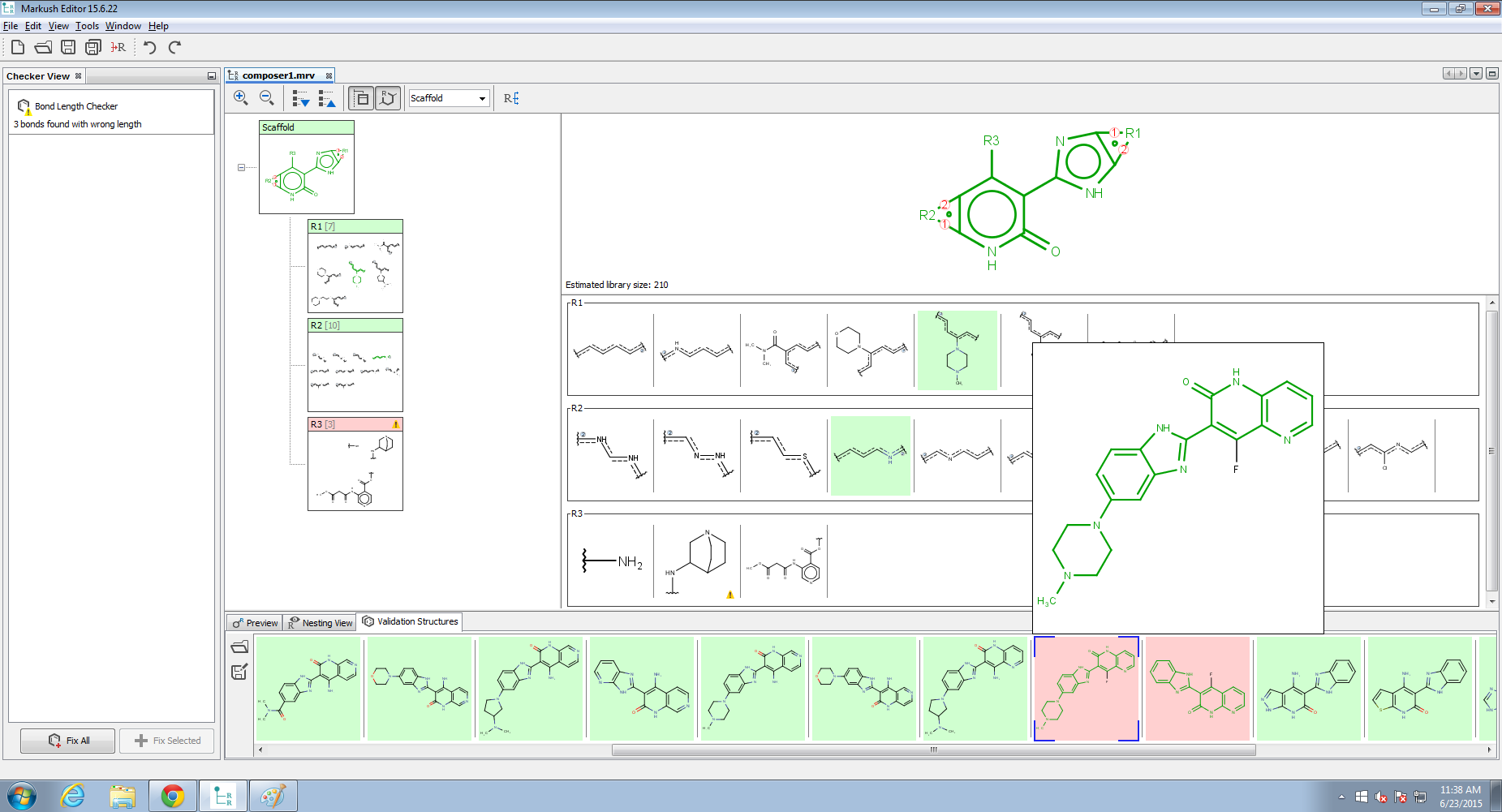
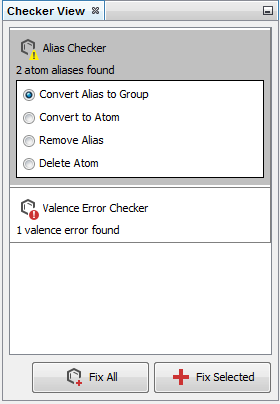
Structure checker panel
Structure checker panel displays the structure drawing errors and warnings related to the active editor component. In the case of an error, an exclamation mark appears in a red circle ![]() , in the case of a warning, a yellow triangle appears
, in the case of a warning, a yellow triangle appears ![]() . By clicking on the checker items, you can choose between the available automatic fixer options. You can fix the issues one-by-one with the Fix Selected button or all together with the Fix All button.
. By clicking on the checker items, you can choose between the available automatic fixer options. You can fix the issues one-by-one with the Fix Selected button or all together with the Fix All button.
Edit Marksuh Structures
Create Markush structure
If you want to create a new Markush structure, click on the New icon,  in the toolbar or select New from the file menu. A new empty scaffold definition appears, and you can draw your scaffold in the previously described way.
in the toolbar or select New from the file menu. A new empty scaffold definition appears, and you can draw your scaffold in the previously described way.
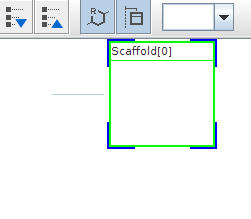
Scaffold can be edited like any R-group definition (see next section). To expand your structure, add R-groups to your scaffold and after saving it, new empty R-group definitions appear and you can continue editing.
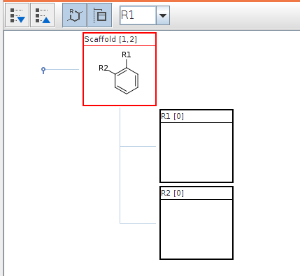
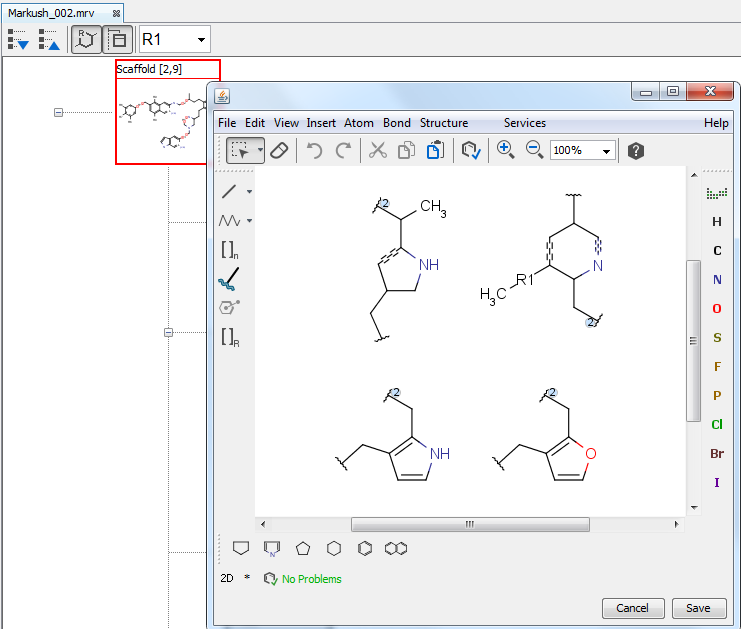
Edit R-groups and fragments
You can edit R-group definitions or individual fragment definitions by right clicking or double clicking on the tree node or the selected fragment. From the context menu, you can select the Edit function or you can add new fragments or remove existing ones.
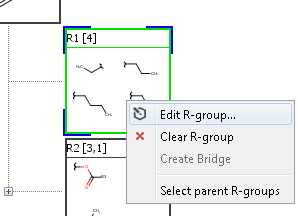
Edit R-group definition from context menu
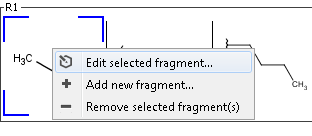
Edit fragment definition from context menu
Sketch window opens with the selected definitions. Modified definitions can be saved by the "Save" button in the bottom right corner of the sketch window, or dropped by pressing the Cancel button.
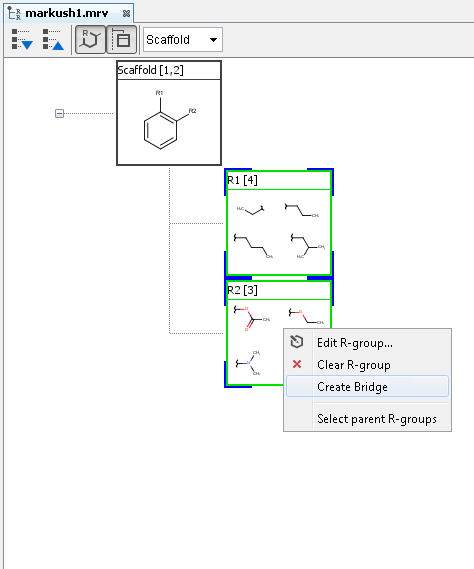
R-group bridging
With R-group bridging, you can create connected R-group definitions. First you need to select two R-groups (on the same level and with one attachment point and only one occurrence in the parent structure) and after right clicking on it, you can select the "Create Bridge" function.
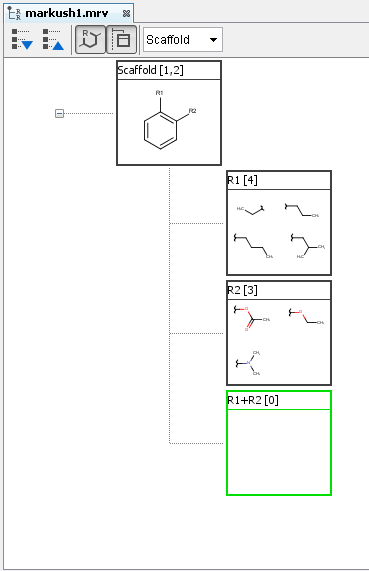
A new empty R-group definition appears with R(x) + R(y) label where x and y denote the number of the originally selected R-groups. You can add fragment definitions with two attachment points to this R-group.
Enumerate Markush Structure
Markush enumeration panel can be opened by clicking on the button ![]() of the toolbar. Enumeration can generate the specific compounds represented by the Markush structure. The following configuration options are available:
of the toolbar. Enumeration can generate the specific compounds represented by the Markush structure. The following configuration options are available:
Main options that specify some essential enumeration settings:
-
Full enumeration: performs exhaustive enumeration of the Markush structure. Markush libraries can potentially be vast in size, so the enumeration is limited to a maximum number of structures that you can specify.
-
Random enumeration: performs random enumeration (sampling) of the Markush structure. This is mostly useful for large Markush libraries where it is not practical to fully enumerate the library. Random enumeration allows you to sample the library in a random fashion so that you obtain a good representation of the various structures in the library. The same warning about the library size that are described for full enumeration also applies to random enumeration.
-
Enumerate homology groups: specifies whether homology groups (Alkyl, Aryl etc.) in the Markush structure should be enumerated with examples of the group. Note that this option gives a representative sample of structures for the homology group, but it is definitely a non-exhaustive enumeration. (Note that homology groups may represent a huge or even infinite number of specific groups.)
-
Number of structures: limits the number of enumerated structures to be generated.
Basic filter options help to eliminate the chemically incorrect enumerated structures:
-
Filter duplicates: eliminate duplicated structures.
-
Filter valance errors: eliminate enumeration of structures with valance errors.
-
Filter infeasible rings: eliminate enumeration of structures with chemically infeasible ring systems.
Display options help to understand the correlation between the enumerated structure and the Markush structure, but they have some performance overhead:
-
Scaffold alignment: preserve the original scaffold orientation in the enumerated structures.
-
Fragment coloring: use different colors to highlight the parts that are derived from different R-groups.
Biased enumeration can only be used with sequential enumeration (currently). Enumerated structures can be filtered according to the predefined property ranges. Biased filters guide the full enumeration process to the desired direction internally, thereby biased enumeration has significant performance benefits compared to post-filtering, but the enumeration process itself can be slower with such biased filers.
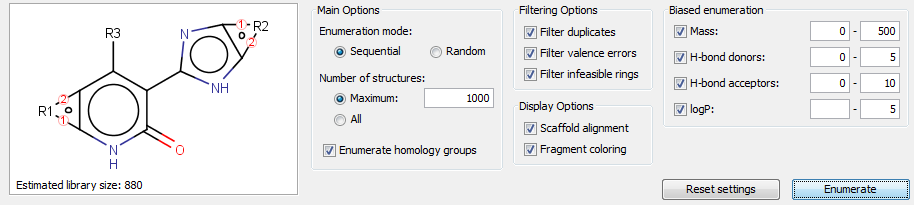
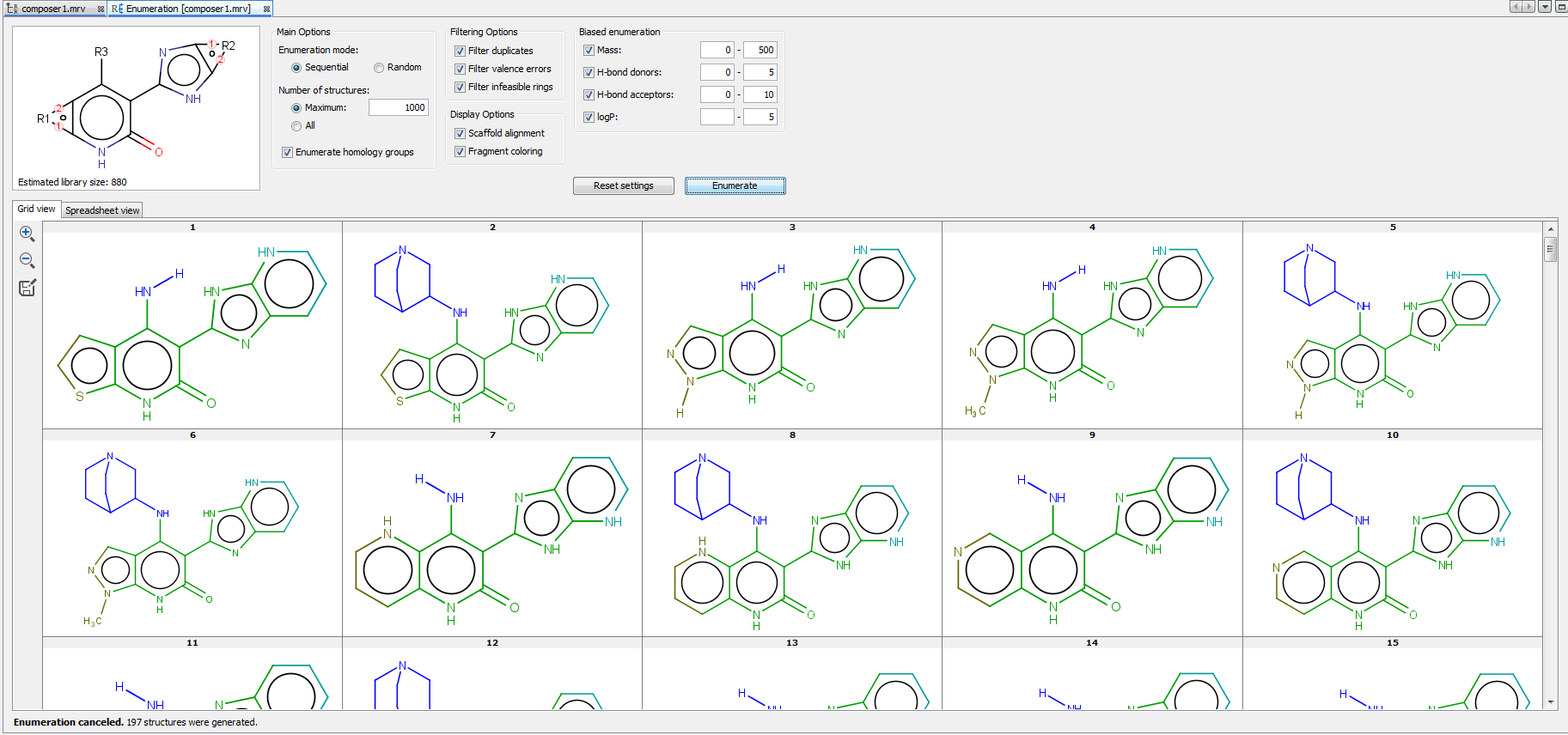
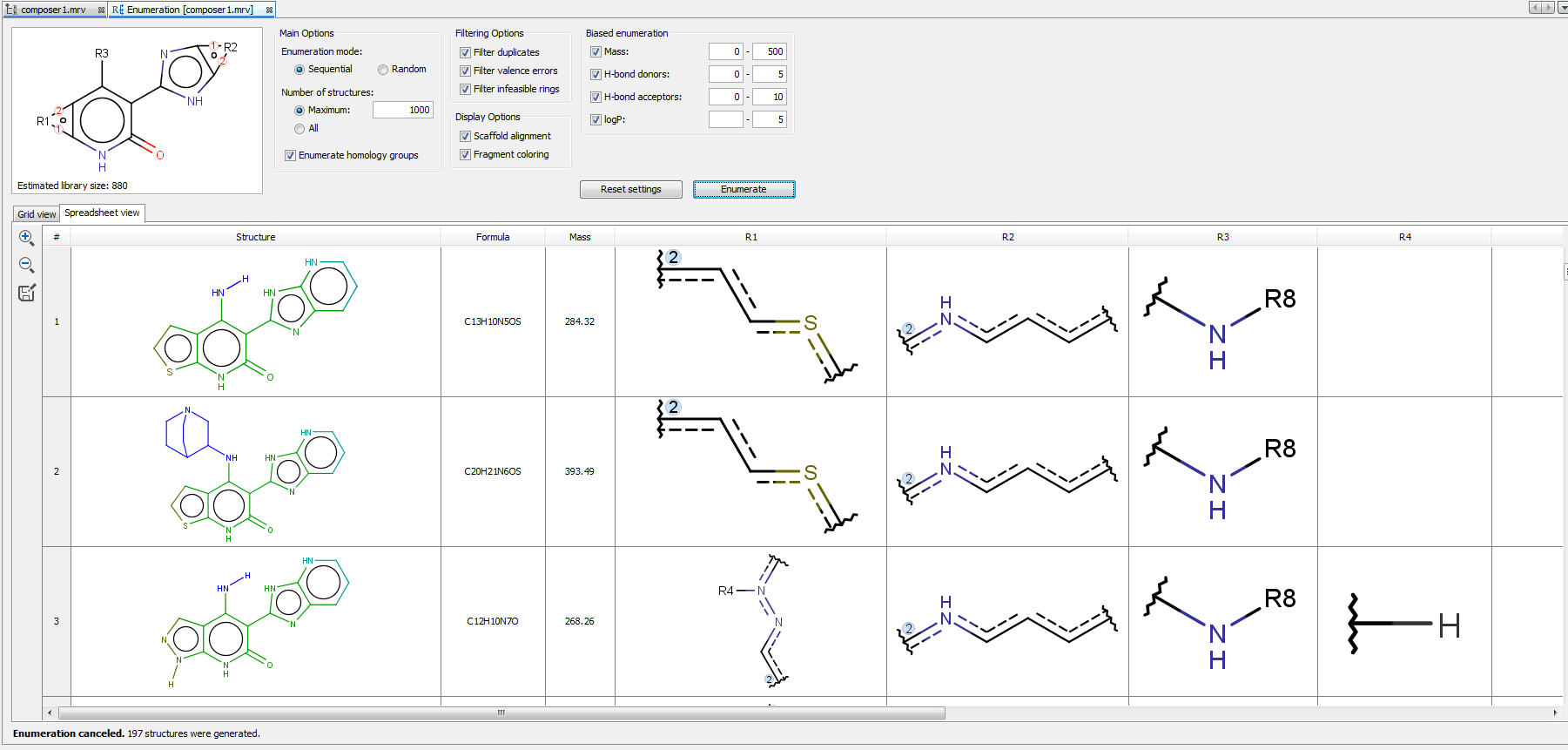
Enumerated structures can be displayed into Grid view and Spreadsheet view. Spreadsheet view contains not only the enumerated structures, but the used R-group fragments and some calculated fields as well, including Formula, Name and Mass. Enumeration results can be saved in various formats by clicking on the icon  in the left sidebar of the enumeration panel.
in the left sidebar of the enumeration panel.
Markush Composer Wizard
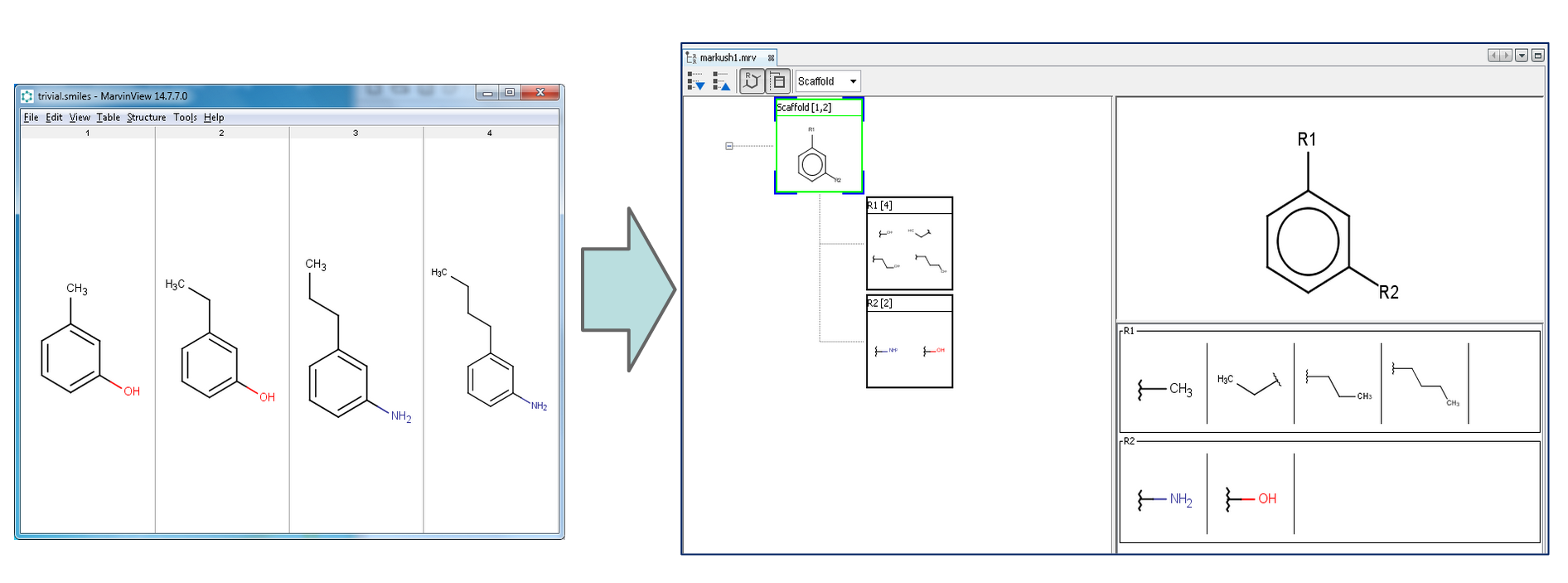
With Markush Composer wizard, you can automatically generate Markush structures from a set of compounds. The compound set has to be in a single file (SDF, SMILES, etc). The composer wizard can be opened from File->Compose Markush Structure...
After selecting this action, the Composer wizard opens.
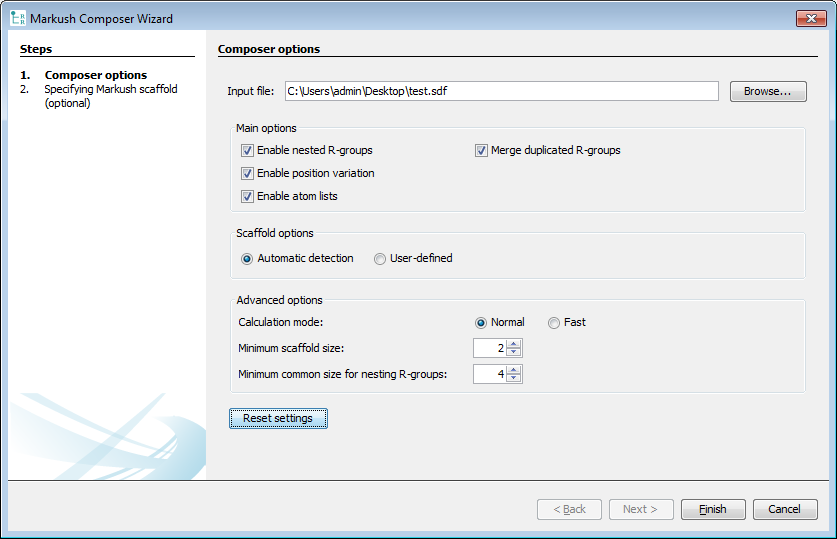
After selecting the Input file , the generation process can be parametrized.
Main options:
If you want to generate single-level Markush structures, uncheck the Enable nested R-groups check box.
If you don't want to reuse R-group definitions, uncheck the Merge duplicated R-groups check box.
If you don't want to generate atom lists, uncheck the Enable atom lists check box.
Scaffold options:
You can choose between Automatic detection of the scaffold or using a User-defined scaffold.
Advanced options:
For processing a very large set of compounds, you can choose a fast search mode instead of normal in Search mode. With fast search mode, the precision of the internal similarity calculations is reduced, which can cause suboptimal resulst in some cases.
If the largest common part of the set of compounds is smaller than the Minimum scaffold size , the composer generates multiple Markush structures with larger scaffolds.
If some of the R-group fragments in an R-group definition have larger common parts than the Minimum common size of nesting R-groups , the algorithm generates a new nested R-group.
If User-defined scaffold option selected, you can navigate to the second page of the wizard dialog with the Next button. In the second page, a preferred scaffold can be set. Input structures not containing the preferred scaffold can be ignored with the Ignore input molecules not containing the scaffold option or in other case composer generates one or more additional Markush structures with different scaffolds for the non-matching input structures. If you do not set any scaffold, Composer tries to find the optimal scaffold automatically. You can use atom lists, bond lists, and R-groups on your predefined scaffold. If you don't define any R-group on this manually specified scaffold, Composer automatically adds R-groups to it, but if you define at least one R-group, Composer uses R-groups exactly how you set them. That is, input molecules with substituents in different position will be ignored.
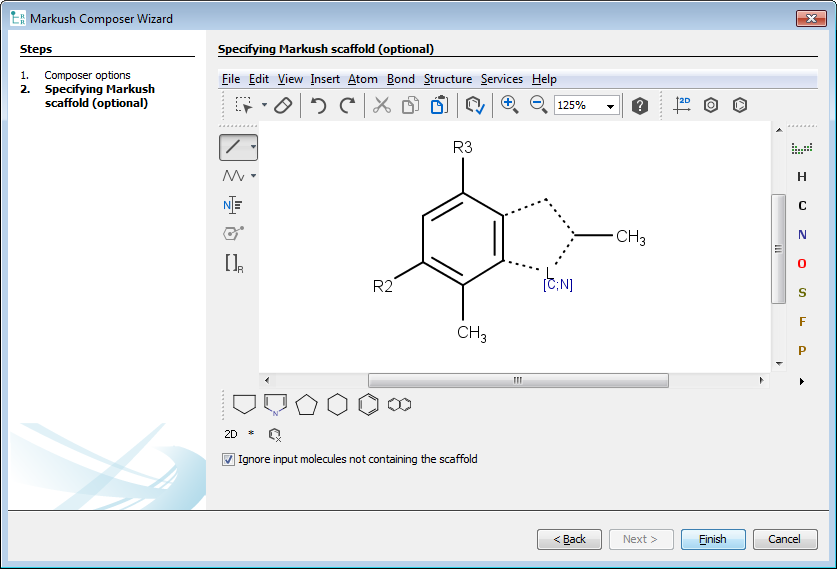
After click on the Finish button, the wizard automatically generates a Markush structure based on the input set.
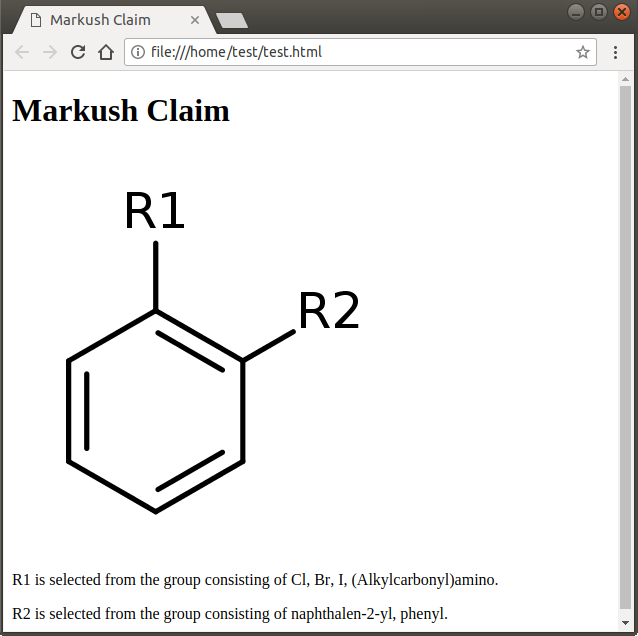
Export Markush structure to text
Markush structure can be exported to text format from the File > Export to Text... menu item. The fragment definitions are converted to text representation and saved as an HTML file. The created file is automatically opened by the default browser on the machine. The content can easily be copied to any word processing application.