Searching in Markush targets tables
Special search types: Markush structures
Contents
A Markush structure is a description of a compound class by generic notations, primarily used in patent claims and the description of combinatorial libraries. The library of a Markush structure is the total set of specific molecules that are described by the Markush structure.
JChem allows searching in combinatorial libraries described as Markush structures, without the need to explicitly enumerate all molecules of the Markush library. The searching can handle the same generic features as the Markush Enumeration Plugin.
Generic Markush features in Markush targets
Currently, JChem supports the following generic features that describe Markush structures in combinatorial libraries:
- R-groups
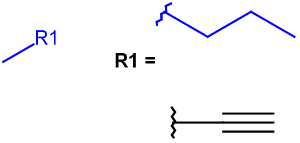
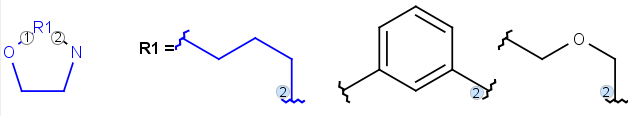
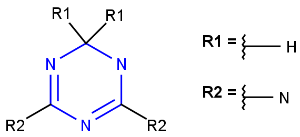
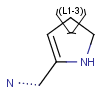
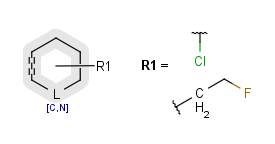
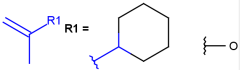
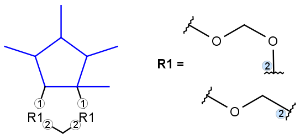
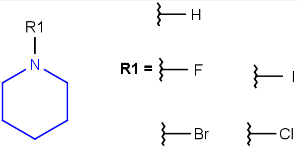
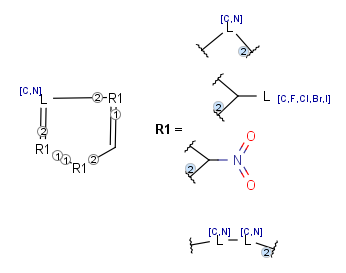
R-groups (also referred to as "substituent variation") are the most widely known Markush generic features. The variable part of the structure is denoted by an R-atom (e.g., R1), and the definitions are given separately. In each definition, the connection points must be defined to show where the bonds of the R-atom are linked. R-atoms can appear in both rings and chains. The same R-atom can appear multiple times, and the different occurrences are handled as different cases. (So they can be substituted with different definitions.) R-logic is not supported. R-group nesting in R-group definitions is allowed to any depth, but without recursion. (An R-group definition cannot use the R-atom it is defining, not even through the use of other embedding R-atom(s).) R-groups up to number R32767 can be used.
| Example | Example Markush library member |
|---|---|
 |
 |
R-group drawing in Marvin Sketch is described in the Marvin Sketch User's Guide.
From version 5.3, R-groups with more than two attachment points are also supported both in searching and in Markush enumeration.
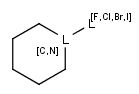
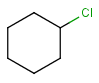
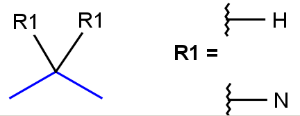
- Atom lists
Atom lists are another example of substituent variation. They define lists of atom types at a given position. There is no restriction for the length of the list and for bond count of atom lists. (Atom list drawing in Marvin Sketch is described here.)
Note : 'Not list'-atoms cannot be present in Markush targets.
| Example | Example Markush library member |
|---|---|
 |
 |
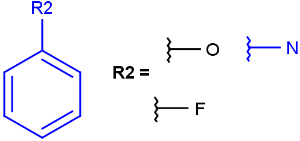
- Bond lists
The following bond lists (generic bond types) are supported: single or double, any (single, double or triple), single or aromatic, double or aromatic. The any bond implicitly can also match aromatic bonds, when it is part of a potentially aromatic system. See: Markush aromatization. In Marvin Sketch, bond lists are accessible amongst query bond types in the bonds pop-up menu.
| Example | Example Markush library member |
|---|---|
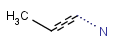 |
 |
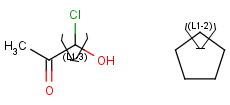
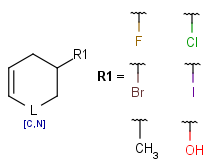
- Link nodes
Link nodes are atoms that may repeat between two of their designated bonds (called outer bonds, denoted by brackets). All other substituents (if exist) repeat together with the atom. In the results, the new bonds between the repeating atoms will have the bond type of the lower order outer bond. Link nodes can be drawn in Marvin Sketch using the popup menu.
| Example | Example Markush library member |
|---|---|
 |
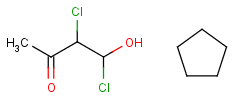 |
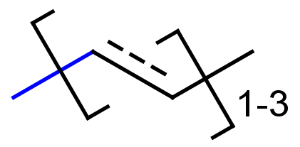
- Repeating units
Repeating units represent structural parts that can be repeated several times. The repeating unit is enclosed in brackets with one or two head and the same number of tail crossing bonds. (Head crossing bonds go through the left bracket.) Two bond pairs represent ladder type repeating units. The repetition range is a comma-separated list of possible repetitions or repetition intervals, e.g. "1,3,5-9". The repetition pattern specifies the way how the subsequent repeated units are linked together: it can be head-to-head(hh), head-to-tail(ht) or either/unknown(eu) (the either/unknown case is not handled by the search software). In case of ladder type repeating units, there is also a flip(f) option that defines that the top and bottom crossing bonds are flipped during each connection. repeating groups with specified repetition ranges.
Substructure search is not yet prepared to handle the case when
- a repeating unit contains another repeating unit or a position variation bond;
- a repeating unit is part of a link node substituent;
- a crossing bond end-atom is an R-atom or a link node;
- repetition range is larger than 500.
Repeating unit drawing is described in the Marvin Sketch Help here, and ladder-type bracket drawing is described at the polymer drawing section.
| Example | Example Markush library members |
|---|---|
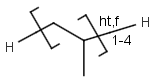 |
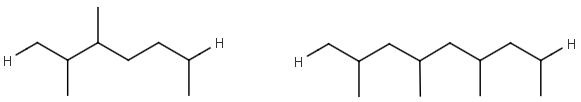 |
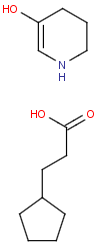
- Position variation bonds
Position variation bonds are bonds attached to variable atoms at one or both end positions. The set of variable atoms is drawn as a multicenter group. A position variation bond connects one atom from one end position to one atom from the other end position. If the end position is a single atom then the bond is attached to this atom, if the end position is a multicenter group then the bond is attached to an arbitrary member of the group. Position variation drawing in Marvin Sketch is described here .
Limitations:
-
Position variation bonds are only allowed to connect atoms of different fragments (i.e., position variation bonds cannot be part of a ring).
-
Multicenter groups are not allowed to contain R-atoms.
-
Multicenter groups are not allowed to contain another position variation bond (i.e., position variation bonds cannot be nested).
If a link node is a member of a multicenter group then the group will also include the repeated atoms if the original multicenter group contains no more atoms from the link fragment, otherwise the position variation bond is part of the link fragment and repeated together with the link node. If the position variation bond is part of the link fragment the multicenter group can have atoms only within the link fragment and the link node atom.
Although an R-atom is not allowed to take part in position variation, it can be the single-atom end position of a position variation bond, in which case its attachment point is connected to the bond.
| Example | Example Markush library members |
|---|---|
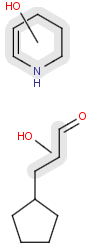 |
 |
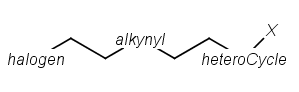
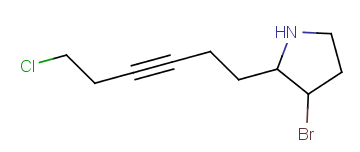
Homology groups stand for sets of homolog molecular parts (e.g., functional groups). These are represented by pseudo atoms labelled with the common chemical annotation of the groups (alkyl, aryl, heterocycle, etc.). See the detailed definition of these groups in a separate document. The pseudo atoms can be most easily drawn in Marvin Sketch using the Homology Groups template group .
| Example | Example Markush library member |
|---|---|
 |
 |
There are two major types of homology groups regarding their way of definition:
-
Built-in groups are defined by specific structural properties of the group. These groups are not enumerated during searching, but the query structure is recognized as fulfilling the requirements for such a structure. The possible number of covered structures is usually infinite, unless the number of atoms is limited. Examples of built-in groups are alkyl, aryl, heterocycle, etc.
-
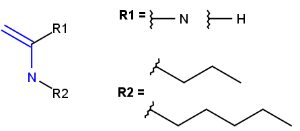
User-defined groups are explicitly defined and only the listed structures can match on these homology groups. The definition is given in the form of an R-group definition, and any of the generic features discussed in this chapter can be used in the definition. These definitions can be customized by the user, and may be context-specific. (E.g. protecting group definition depends on which functional group it is protecting.)
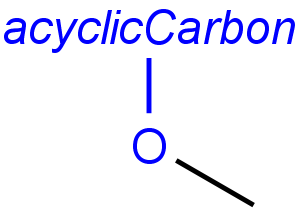
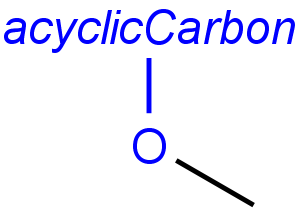
Handling of target side homology groups is controlled by the homology broad translation option. If matching is switched off for a query atom, then this query atom can match homology atom only if it is the same homology atom. If matching is switched on for a query atom, it matches homology group representing a larger set of structures. E.g. acyclic carbon atom can match alkyl or carbontree, an Fe atom can match transition metal or metal homology atom. Homology atoms can also match homology atoms covering a larger set of structures: carboalicyclyl can match cyclyl or xx. In these cases, the query homology atom is a subset of the target homology atom. Subset rules can be seen here. Possible values for homology translation:
all
all query atoms can match on broader homology atoms, if the properties of the group are fulfilled.
none
no query atoms can match on broader groups, they can only match if the query atom is a homology atom of the same group.
marked
only the marked query atoms can match on broader homology groups. This option is not yet implemented, it works as none.
The default value for homology translation is none.
Translation NONE behavior
-
homology groups cannot match and cannot be matched by specific atoms or homology groups being a superset or a subset of the given group.
-
homology groups can match pseudo atoms with alias name of the given group (e.g., chk matches alkyl)
-
homology properties are ignored (e.g., alkyl,LO matches alkyl,HI)
Read more about homology groups.
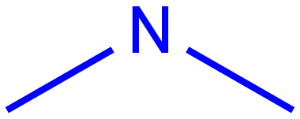
- Query atoms
In case of Markush search, some query atoms can be used on the target side. They can be matched by the same query atom or by specific atoms that can be the hit of the given query atom. Targets containing only query atoms beside specific structures can be searched by query side homology groups. Query atoms are not enumerated with the Markush enumeration functionality.
Query atoms supported on the target side: A, AH, Q, QH, M, MH, X, XH, G<n>.
Querying Markush targets
The following search types are allowed for Markush targets/tables: DUPLICATE, SUBSTRUCTURE, FULL and FULL_FRAGMENT search.
SUPERSTRUCTURE search is not allowed for targets in Markush tables, however, it is allowed for Markush targets in files (in memory search). SUPERSTRUCTURE search for Markush targets is allowed in Query tables.
Similarity search is not allowed for Markush targets.
DUPLICATE, FULL and FULL_FRAGMENT search cannot be combined with tautomer search in case of Markush targets/tables.
Duplicate search can be used to check if the same structure is inserted to a database table again. Note that it requires entire drawing equality for matching, it does not check Markush overlap: it does not give hit if two markushes are different, but their sets of represented specific structures are the same.
The following query features are supported in the query when searching Markush targets:
-
Query atoms, including atom lists, not lists, generic query atoms (A, Q, M, etc.). Examples
-
Atom query properties: a (aromatic), A (aliphatic), R (part of a ring), R0 (not part of a ring), R<n> (number of rings the atom is member of), s (substitution as drawn), s<n> (exact substitution count), D<n> (number of explicit connections), v<n> (valence), rb (ring bond count as drawn), rb<n> (exact ring bond count), u (unsaturated bonds), X<n> (number of connections), H<n> (number of hydrogen substituents). Examples
-
Query bond types, including any, single/aromatic, etc.
Query Markush Hit 
-
Bond topology query properties (chain/ring) on bonds
Query Markush Hit 
-
Tetrahedral and double bond E/Z stereochemistry
Query Markush Hit 
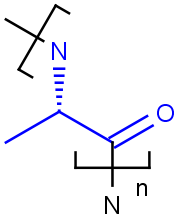
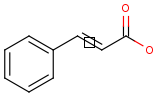
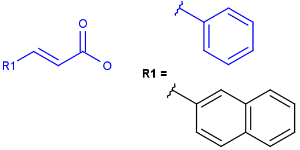
-
Link nodes
Query Markush Hit 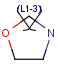
-
Position variation bonds
Query Markush Hit 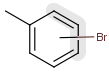
-
Explicit H atoms in query match both explicit and implicit H atoms in target. Attention: in case of full search, explicit hydrogens are not considered in R-groups or atom lists. In the future, matching of Hydrogens will be further improved.
Query Markush Hit 
-
Simple R-group queries are supported when searching Markush targets. The R-group query is a Simple R-group query if:
-
it does not have R-logic;
-
number of enumerates does not exceed 100.
-
R-group queries of Markush targets are not supported with
undefinedRAtom:g/gh/gheoptions when query structures contain undefined R-atom(s). They are supported only withundefinedRAtom:aandundefinedRAtom:uoptions.Query Markush Hit 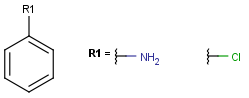
-
-
Homology groups on target side (and on query side) under the following conditions:
Examples
Table 1. Simple substructure search examples (the bond denoted by dots is an "any" bond)
| target | ||||
|---|---|---|---|---|
 |
 |
 |
||
| substructure query |  |
 |
 |
 |
 |
 |
 |
 |
|
 |
 |
 |
 |
Table 2. Simple full structure search examples (the bond denoted by dots is an "any" bond)
| target | ||||
|---|---|---|---|---|
 |
 |
 |
||
| full structure query | 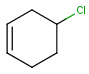 |
 |
 |
 |
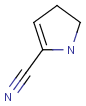 |
 |
 |
 |
|
 |
 |
 |
 |
Table 3. Accepted query atoms on the query side in Markush search
| Option | Query Structure | Markush Target Hit |
|---|---|---|
| atom list | 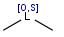 |
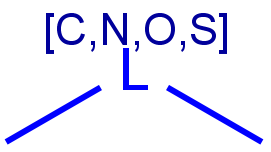 |
| not list |  |
 |
| Aany atom except H | 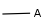 |
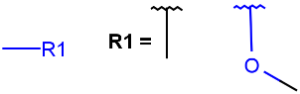 |
| AHany atom including H(temporarily only explicit H) | 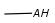 |
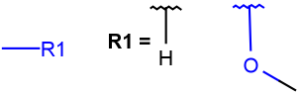 |
| Qany atom except C and H |  |
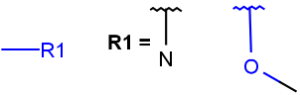 |
| QHany atom except C including H(temporarily only explicit H) | 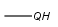 |
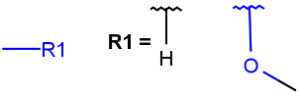 |
| M any metal |  |
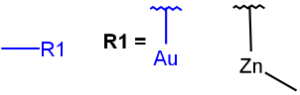 |
| MHany metal or H (temporarily only explicit H) | 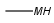 |
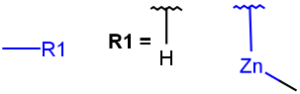 |
| Xany halogen |  |
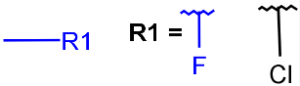 |
Table 4. Accepted atom properties on the query side in Markush search
| Property | Query Structure | Markush Target Hit |
|---|---|---|
| aaromatic | 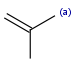 |
 |
| Aaliphatic | 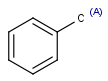 |
 |
| Rpart of a ring | 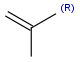 |
 |
| R0not part of a ring | 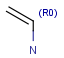 |
 |
| R<n>number of rings the atom is member of | 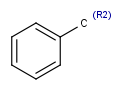 |
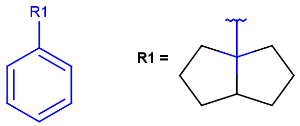 |
| s*substitution as drawn(including its substituents in the query structure) | 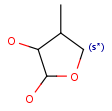 |
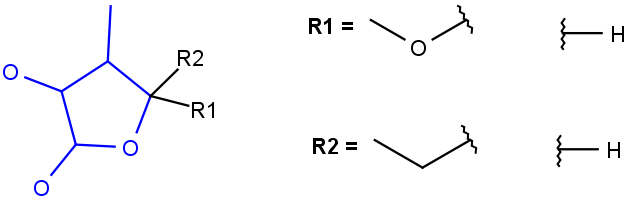 R1 and R2 must be H in the hit R1 and R2 must be H in the hit |
| s<n>exact substitution count(including its substituents in the query structure) | 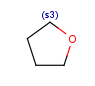 |
 R1 or R2 must be H in the hits R1 or R2 must be H in the hits |
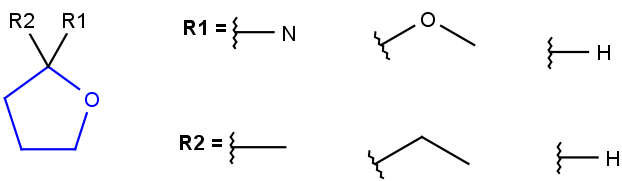
| D<n>number of explicit connections(equivalent to s<n>) |  |
 R1 or R2 must be H in the hits R1 or R2 must be H in the hits |
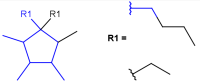
| v<n>valence | 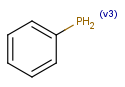 |
 |
| rb*ring bond count as drawn | 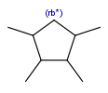 |
 |
| rb<n>exact ring bond count | 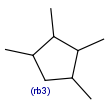 |
 |
| uunsaturated bonds |  |
 |
| X<n>connections |  |
 |
| H<n>number of hydrogen substituents |  |
 |
Markush structure reduction to a hit
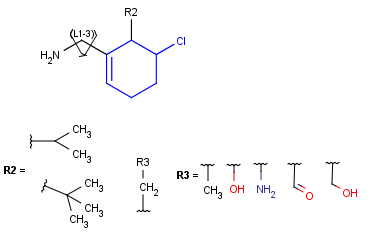
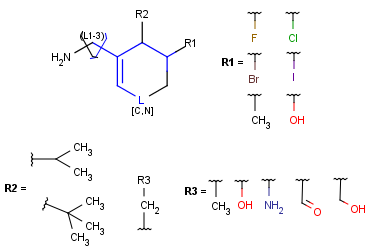
When a query matches a Markush structure, there are different ways of displaying the hit. One possibility is to color the matching parts of the original Markush structure, but it may mean that the highlighting is spread across different fragments (R-group definitions) when the query overlaps variable parts. Markush structure reduction is a technique wherein the variable parts overlapping the hit are expanded (substituted with the appropriate specific definition). This way the hit highlighting is always visible as a whole and part of the scaffold. (Note that the resulting structure of Markush structure reduction may still contain generic features.)
Markush structure reduction examples
Table 5. Reduction of Markush Structure
| target | |||
|---|---|---|---|
| Hit coloring in original Markush structure | Markush structure reduction to the hit | ||
| substructure query |  |
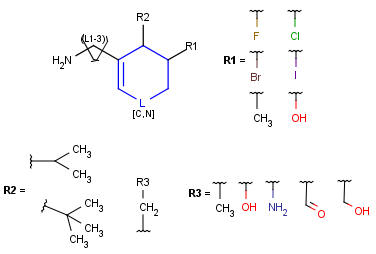 |
 |
 |
 |
 |
Markush aromatization
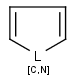
With the introduction of generic notation in target structures, it is possible to formulate ring systems with ambiguous aromaticity status: some enumerations of the ring are aromatic, and others are not. See a simple example below.
 |
|---|
Therefore, in case of Markush targets, it is not possible to entirely separate standardization and searching the way as described in section Standardization JCB. Instead, aromaticity is handled in a more complex way that ensures that no matching is lost. (However, there may be false positives in case the query is not matching a full ring. See examples below.)
Standardization for Markush targets (tables) solely consists of a special aromatization method: Markush aromatization. It divides rings of the Markush structure with generic features into three different categories:
-
aromatic (the ring describes only aromatic rings)
-
non-aromatic (the ring describes only non-aromatic rings)
-
ambiguous (the ring describes both aromatic and non-aromatic rings) (Too complex rings that cannot be decided stay in the ambiguous category. Currently, the default complexity limit is 100 enumerations of the generic features causing ambiguity.)
Searching considers aromatic and non-aromatic rings the same way as for specific structures. However, ambiguous rings are allowed to be matched by both aromatic and non-aromatic query parts.
Examples
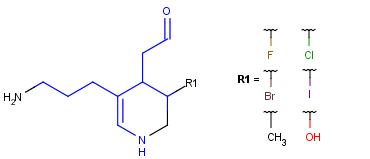
Table 6. Aromaticity in Markush targets
| target | |||
|---|---|---|---|
 |
 |
||
| substructure query | 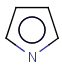 |
 |
 |
 |
 |
 |
|
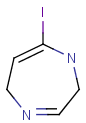 |
 |
 |
|
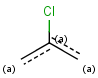 |
 |
 |
Creation of R-group definitions from a structure file
If you want to use a file of molecules (such as e.g. acyl-halide reagent library) as R-group definitions in Markush structures, you can transform the file by using R-group Decomposition or Reagent Clipping.
-
R-group decomposition is a special kind of substructure investigation that aims at finding a central structure and identifying its ligands at certain attachment positions. The query molecule consists of the scaffold and ligand attachment points represented by R-groups. After specifying the query structure, the process filters and identifies ligands in the target library, tabulates decomposed R-groups, and creates Markush structure from R-table. See details on making R-group definitions by R-group decomposition.
Figure 1. Progress of R-group decomposition
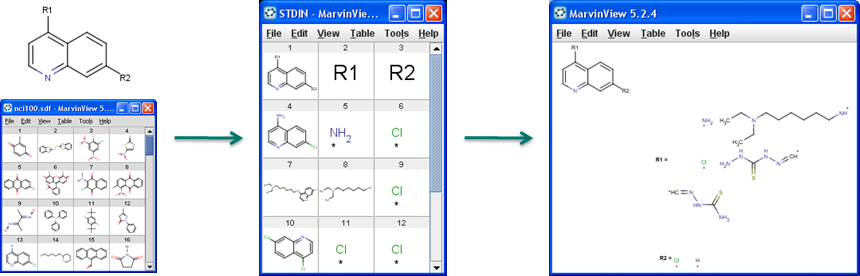
-
Reagent Clipping
-
Clip reagent group and replace by attachment point: Reactor and Standardizer can handle transformations that includes the creation of attachment points. This way the reacting group can be replaced by an attachment point by a transformation like:
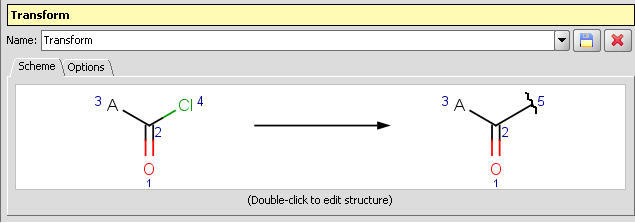
-
Merge the above transformed reagents into one diagram so that each record will be a separate R-group definition. This is already available using the molconvert command-line program, with option -R (read about the usage of MolConverter)
Example:
molconvert mrv scaffold.mrv -R1 r1_definitions.mrv -R2 r2_definitions.mrv
-