Local Services
Introduction
Local services are used for accessing third-party calculations implemented in Java. They do not require a server or network connection. A Java Archive (JAR) file acts as the server, any public class with default constructor acts as a service, and all public methods can be called. Please keep in mind that the build-in editor may hide methods that require unsupported argument types. Direct access of the Marvin Services API does not have this restriction, any type can be used.
This is the easiest way to embed third-party calculation to the MarvinSketch application, cxcalc, or Chemical Terms. However, java coding is required to assemble the JAR files. Also, note that these services cannot be accessed from a non-java environment such as Marvin .NET or JChem for Excel.
{primary} Classes used through service calls must be stateless, as each service call creates a new instance of the class by the default constructor before calling the function.
Examples
Example 1
In this basic example, we implement a method that returns the string "Hello <molecule name>!" , where <molecule name> is the name of the molecule generated by ChemAxon's S2N (Structure to Name) converting tool. The following code will do this:
package chemaxon.examples.calcintegration;
import chemaxon.struc.Molecule;
public class Hello {
public String helloMolecule(Molecule mol) {
return "Hello "+mol.toFormat("name")+"!";
}
}-
Having compiled this code, pack the created class files into a JAR file. All Java IDEs can export Java files in a JAR file. In Eclipse, it can be done through the File > Export menu item.
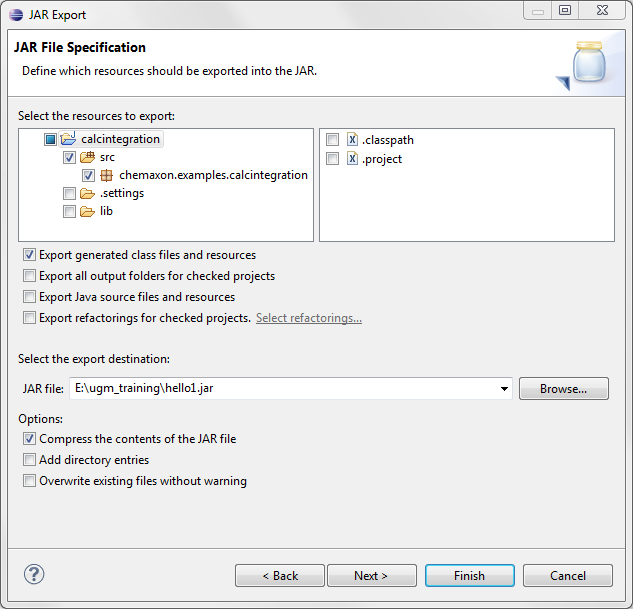
-
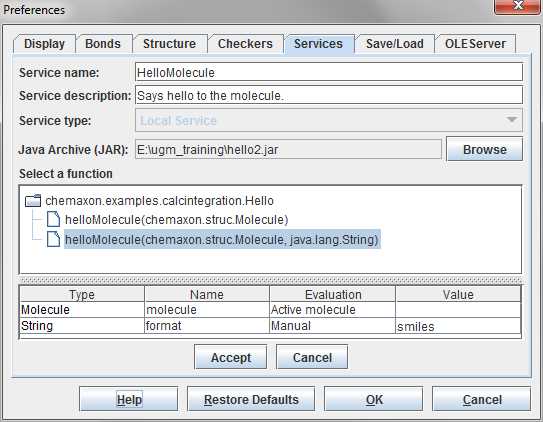
After creating the JAR file, open MarvinSketch. Please note that version 5.6 or later is required. Select Edit > Preferences > Services from the menu. Then press the + button to add a new service.
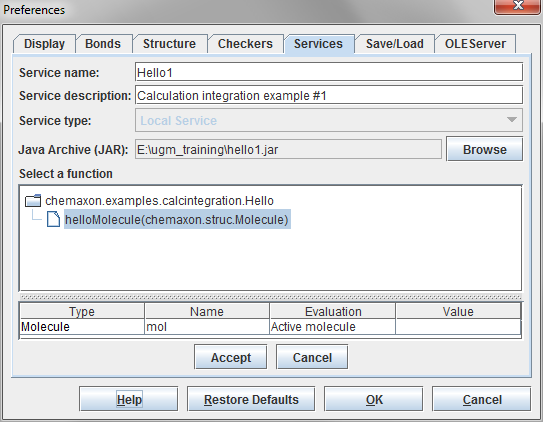
-
To add a new service then proceed to press the Accept button. It will appear in the list of services.
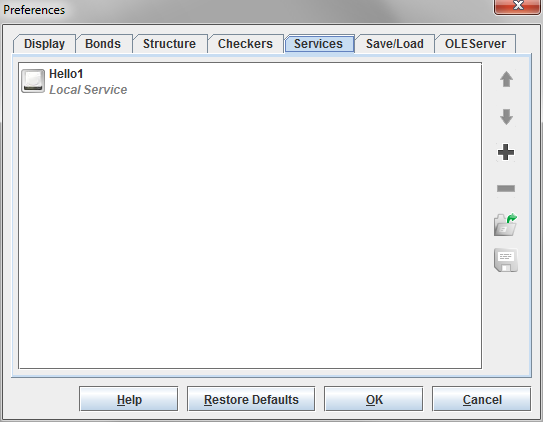
-
As a result of these steps, the new service is available from the Tools > Services menu.
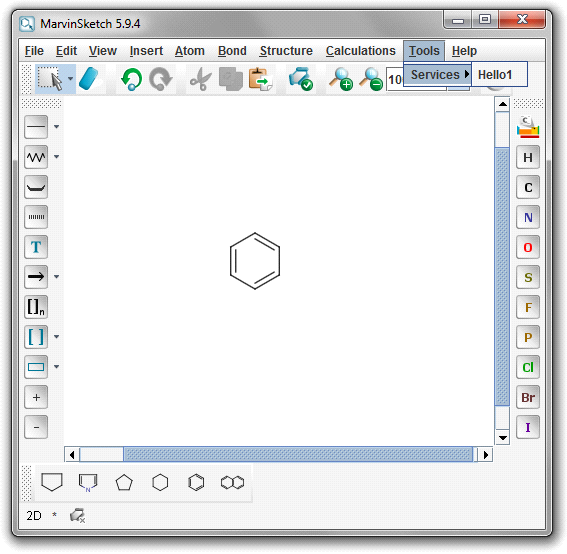
-
Finally, to use the new service, select the Tools > Services > Hello1 menu item with an input molecule on the canvas. The calculation result appears in a new dialog.
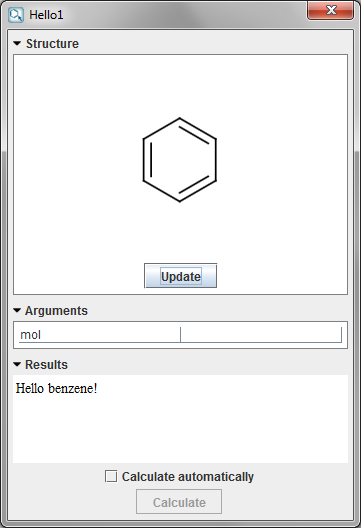
{info} Local Service makes good use of the Alias and Description annotations. Any method annotated can provide default names and descriptions for services and arguments. Also, these aliases are available from cxcalc as well, so a default service and argument name can be guaranteed by the class author.
Now let's extend this example by adding a new method to the Hello.java class and adding Alias and Description annotations to it. Thus the new code will be:
@Alias(name="HelloMolecule", params={"molecule", "format"})
@Description("Says hello to the molecule.")
public String helloMolecule(Molecule mol, String format) {
return "Hello "+mol.toFormat(format)+"!";
} -
Create a JAR file as above and name it hello2.jar. Call the new method (as a local service) from this JAR file. The name of the service, the description, and the parameter names will be automatically filled during the setup.
-
A result example in this case is:
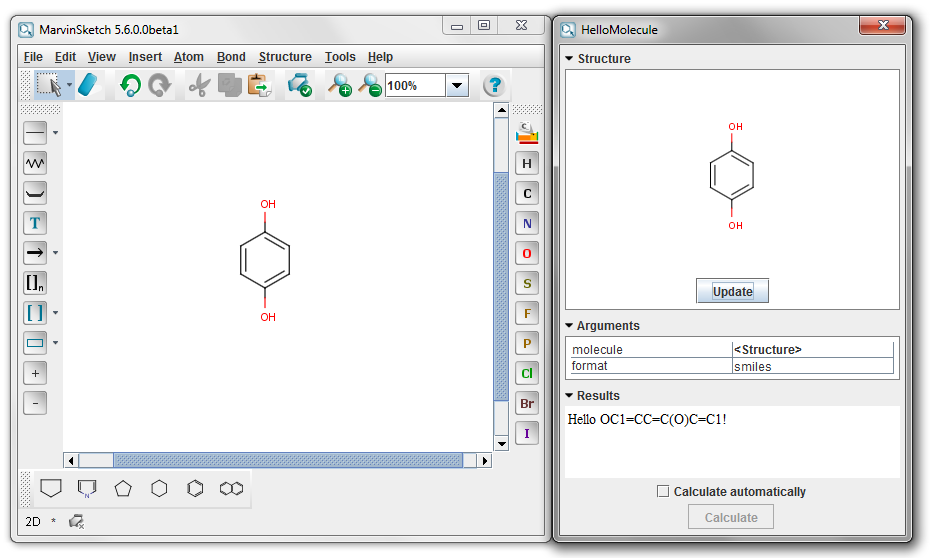
{info} The result molecule format can be modified on the result display panel.
Example 2
The next example describes the implementation of some microspecies related calculations.
It calculates the major microspecies at a given pH, the microspecies count at a given pH, and generates an HTML report about the calculations.
package chemaxon.examples.calcintegration;
import java.io.File;
import java.io.FileOutputStream;
import java.io.IOException;
import chemaxon.marvin.calculations.MajorMicrospeciesPlugin;
import chemaxon.marvin.plugin.PluginException;
import chemaxon.marvin.services.localservice.Alias;
import chemaxon.marvin.services.localservice.Description;
import chemaxon.struc.Molecule;
public class MicrospeciesCalculator {
private static MajorMicrospeciesPlugin createPlugin(Molecule mol, Double pH) {
MajorMicrospeciesPlugin mmsp = new MajorMicrospeciesPlugin();
boolean valid = false;
try {
mmsp.setMolecule(mol);
mmsp.setpH(pH);
valid = mmsp.run();
} catch (PluginException e) {
// error, valid == false
}
return valid ? mmsp : null;
}
@Alias(name="MMS", params={"mol", "pH"})
@Description("Calculates major microspecies of the given " +
"molecule at given pH.")
public Molecule getMajorMicrospecies(Molecule mol, Double pH)
throws PluginException {
MajorMicrospeciesPlugin plugin = createPlugin(mol, pH);
return plugin == null ? null : plugin.getMajorMicrospecies();
}
@Alias(name="MSCount", params={"mol", "pH"})
@Description("Counts microspecies of the given molecule at " +
"given pH.")
public Integer getMicrospeciesCount(Molecule mol, Double pH)
throws PluginException {
MajorMicrospeciesPlugin plugin = createPlugin(mol, pH);
return plugin == null ? null : plugin.getMicrospeciesCount();
}
@Alias(name="MSReport",
params={"mol", "pHLower", "pHUpper", "displayImage"})
@Description("Creates HTML report about microspecies.")
public String getMicrospeciesHtmlReport(Molecule mol,
Double pHLower, Double pHUpper, Boolean displayImage)
throws PluginException, IOException {
MajorMicrospeciesPlugin plugin = createPlugin(mol, pHLower);
if (plugin == null) {
return null;
}
// calculate, build and return the result string
StringBuilder rbuilder = new StringBuilder("<html><body>");
rbuilder.append("<table border=\"1\">");
rbuilder.append("<tr>");
rbuilder.append("<th><b>pH</b></th>");
rbuilder.append("<th><b>Major microspecies</b></th>");
rbuilder.append("<th><b>Charge</b></th>");
rbuilder.append("</tr>");
for (int i=0;i<=pHUpper.intValue()-pHLower.intValue();i++) {
double pH = pHLower+i;
plugin.setpH(pH);
plugin.run();
Molecule mms = plugin.getMajorMicrospecies();
rbuilder.append("<tr>");
rbuilder.append("<td>"+pH+"</td>");
if (displayImage) {
File f = File.createTempFile("image"+i, "png");
FileOutputStream fos = new FileOutputStream(f);
fos.write(mms.toBinFormat("png"));
rbuilder.append("<td><img src=\""
+f.toURI()+"\"></td>");
} else {
rbuilder.append("<td><font color='blue'><i>"
+mms.toFormat("smiles")+"</i></font></td>");
}
rbuilder.append("<td>"
+mms.getFormalCharge()+"</b></td>");
rbuilder.append("</tr>");
}
rbuilder.append("</body></html>");
return rbuilder.toString();
}
}-
Pack the MicrospeciesCalculator class into a JAR file as already described ( mscalc.jar ). Add all methods as local services to the service list.
-
The result of running the MMS calculation is the major microspecies molecule at a given pH.
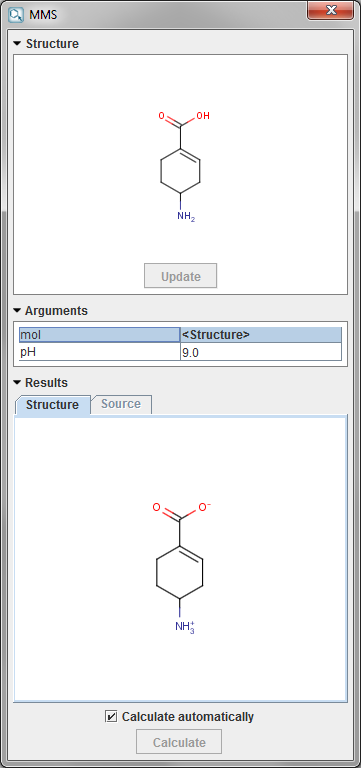
-
The MSReport calculation generates the source code of an HTML page and returns it as a string. The HTML page is then rendered on the fly by the result display component.
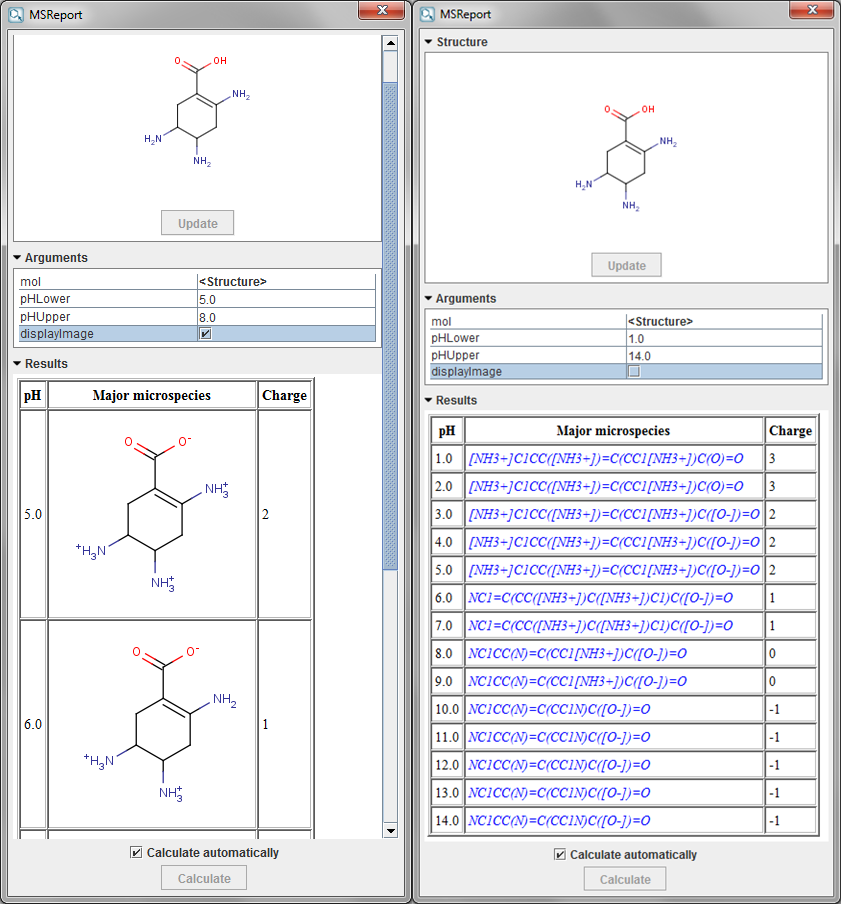
Changes made on the molecule in MarvinSketch will be automatically updated on the results display (both structure and results) if the Calculate Automatically option is enabled.
Calling Local Services from API
The following code snippet calls the Integer countAtoms(Molecule) function of the example.services.SampleService class located in localserviceexample.jar.
// input molecule
Molecule input = MolImporter.importMol("c1ccncc1");
// initialize descriptor
LocalServiceDescriptor descriptor = new LocalServiceDescriptor();
descriptor.setURL("/path/to/localserviceexample.jar");
descriptor.setClassName("example.services.SampleService");
descriptor.setMethodName("countAtoms");
descriptor.addArgument(ServiceArgument.createArgument(new Molecule()));
// asynchronous call
descriptor.getServiceHandler().callService(descriptor, new AsyncCallback<Integer>() {
@Override
public void onSuccess(Integer result) {
System.out.println("Asynchronous call returned " + result);
}
@Override
public void onFailure(ServiceException caught) {
System.err.println("Asynchronous call failed.");
}
}, input);
// synchronized call
Object result = null;
try {
result = descriptor.getServiceHandler().callService(descriptor, input);
} catch (ServiceException e) {
System.err.println("Service call failed.");
}
System.out.println("Synchronized call returned " + result);
Use Annotations to Define Default Names and Description
A Local Service can look up default service and argument names, as well as description information from annotations. These values are used in MarvinSketch when adding the Local Service to the list of services by automatically completing the form. The values can be edited manually, but the defaults are always available from Chemical Terms or cxcalc - as well as the optionally overwritten ones.
You can find a sample class that can be used as a Local Service below. To download the sample service jar file with the source, click here.
*
* Copyright (c) 1998-2014 ChemAxon Ltd. All Rights Reserved.
*
* This software is the confidential and proprietary information of
* ChemAxon. You shall not disclose such Confidential Information
* and shall use it only in accordance with the terms of the agreements
* you entered into with ChemAxon.
*
*/
package example.services;
import chemaxon.formats.MolFormatException;
import chemaxon.formats.MolImporter;
import chemaxon.marvin.services.localservice.Alias;
import chemaxon.marvin.services.localservice.Description;
import chemaxon.struc.Molecule;
/**
* This is a sample class to demonstrate how to write
* classes for Marvin Services Local Service implementation.
* @author Istvan Rabel
*/
public class SampleService {
/**
* Returns the number of atoms in the specified molecule
* @param molecule the molecule being checked
* @return the number of atoms in the molecule
*/
/*
* (non-javadoc)
* This method can be called as a LocalService from
* Marvin Sketch, cxcalc and Chemical Terms.
* Annotations are used to provide default names
* for Service and arguments, as well as a description.
*/
@Alias(name="AtomCount", params={"Structure"})
@Description("Returns the number of atoms in the structure")
public Integer countAtoms(Molecule molecule) {
return molecule.getAtomCount();
}
/**
* Returns a formatted (HTML) message with the number of
* atoms in the molecule imported from argument.
* @param moleculeString a string representation of a molecule
* @return a formatted (HTML) message with the number of atoms
*/
/*
* (non-javadoc)
* This method can be called as a LocalService from
* Marvin Sketch, cxcalc and Chemical Terms.
* Annotations are used to provide default names
* for Service and arguments, as well as a description.
*/
@Alias(name="AtomCountText", params={"Molecule"})
@Description("Returns a formatted text message containing the number of atoms in the structure.")
public String countAtomsHTML(String moleculeString) {
// import the molecule
Molecule molecule = null;
try {
molecule = MolImporter.importMol(moleculeString);
} catch (MolFormatException e) {
// invalid molecule string
molecule = new Molecule();
}
// get the atom count
int value = countAtoms(molecule);
// build and return the result string
StringBuilder builder = new StringBuilder("<html><body>");
if(value > 1) {
builder.append("The structure has <font color='blue'><b>");
builder.append(value);
builder.append("</b></font> atoms.");
} else {
builder.append("The structure has <font color='red'><i>"
+ (value == 0 ? "no atoms" : "only one atom")
+ "</i></font>.");
}
builder.append("</body></html>");
return builder.toString();
}
}