Homology Groups in Markush Structures
Currently JChem enables the searching of Markush structures containing Homology groups only with specific molecule queries (with no query features). Homology groups are supported on the target and on the query side (the latter only for non-Markush targets). Properties can also be specified for the groups.
Read the user's guide about Homology groups and editing their properties in MarvinSketch and in Marvin JS.
Contents
Definition of Homology groups
Homology groups are represented by Pseudo atoms, labeled with common chemical annotations of these groups. Most groups have Alias names that allow shorter names. The names are case insensitive, spaces might be inserted.
Pseudo atoms can be easily drawn in Marvin Sketch using the Homology groups template group.
There are two major types of Homology groups regarding the way of their definition:
-
Built-in Homology groups are defined by specific structural properties of the group. These groups are not enumerated during the search, but the query structure is recognized when fulfills the requirements for such a structure. The possible number of covered structures is usually infinite, unless the number of atoms is limited. Examples of built-in groups are alkyl, aryl, heterocycle, etc.
-
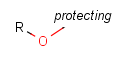
User-defined Homology groups are explicitly defined and only the listed structures can match these homology groups. The definition is given in the form of an R-group definition and any of the generic features (discussed in the Markush chapter) can be used in the definition. There are some Predefined and new 'User-defined' groups can be added, too. These 'User-defined' definitions can be customized by the user, and they can be context-specific. (E.g. Protecting group definition will depend on the functional group, which is protected.)
1. Built-in Homology groups
Table 1. shows the properties of the Built-in Homology groups. The groups are recognized having specific features. These features are shown in the table as "compulsory" parts. Optional parts are structures that are not necessarily part of the structure that matches on the Homology group.
'Incomplete case' means structures that are substructures of structures that would represent a homology group. It is required that even such incomplete structures should be extended to a complete homology group. Search options regulate if incomplete structures can match on a homology group or complete structures are required.
The "Example" column shows complete structures representing the homology groups.
Table 1. Built-in Homology groups
| Group name (Alias names) | Compulsory | Optional | Example |
|---|---|---|---|
| Alkyl (CHK) | - minimum of one carbon atom - only carbon and hydrogen atoms - single bonds - no ring bonds | connection point at arbitrary position(s) | 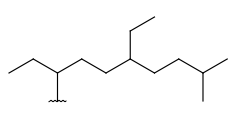 |
| Alkenyl (CHE) | - at least one double bond - minimum of 2 carbon atoms - otherwise same as for Alkyl | same as above | 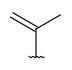 |
| Alkynyl (CHY) | - at least one triple bond - minimum of 2 carbon atoms - otherwise same as for Alkyl | same as above, double bond | 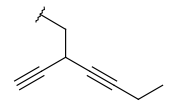 |
| CarbonTree (acyclicCarbon) | Any connected acyclic carbon structure. | - | 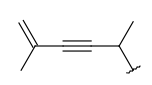 |
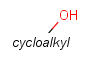
| Carboalicyclyl (CYC, cycloalkyl) | - monocyclic or fused aliphatic rings - only carbon and hydrogen atoms - no substitution by (saturated) alkyl chains | - double or triple bonds in the ring but not aromatic - several connection points on the rings | 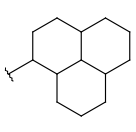 |
| Aryl(since version 17.21.0) | - monocyclic or fused rings- among these rings at least one should be aromatic | - double bonds/triple bonds in the aliphatic rings- several connection points but all must be on an aromatic ring (can't have external connection on an aliphatic ring) | 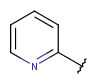 |
| Carboaryl (ARY) | - monocyclic or fused rings - among these rings at least one should be aromatic - only carbon and hydrogen atoms | - double bonds/triple bonds in the aliphatic rings - several connection points but all must be on an aromatic ring (can't have external connection on an aliphatic ring) | 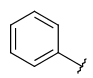 |
| Heteroalicyclyl (AliphaticHeterocyclyl)(since version 17.21.0) | - monocyclic or fused aliphatic rings with at least one hetero atom; carbon atom is also required | same as carboalicyclyl | 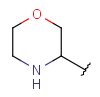 |
| Heteromonoalicyclyl (HET) | - monocyclic aliphatic ring with at least one hetero atom; carbon atom is also required | same as carboalicyclyl | 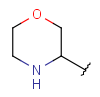 |
| Fusedheteroalicyclyl (Heteropolyalicyclyl)(since version 17.21.0) | - fused aliphatic rings with at least one hetero atom; carbon atom is also required | same as carboalicyclyl | 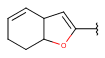 |
| Heteroaryl(since version 17.21.0) | - monocyclic or fused aromatic rings with at least one hetero atom, carbon atom is also required | same as aryl | 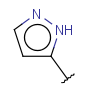 |
| Heteromonoaryl (HEA) | - similar to aryl but the monocyclic aromatic ring should contain at least one hetero atom, carbon atom is also required - no fused rings | same as aryl | 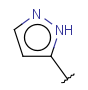 |
| Fusedheteroaryl (Heteropolyaryl)(since version 17.21.0) | - fused aromatic rings with at least one hetero atom; carbon atom is also required | same as aryl | 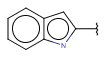 |
| FusedHeterocyclyl (HEF, fusedHetero) | - Fused rings having at least one hetero atom, carbon atom is also required | same as aryl, but the connection point can be on an aliphatic ring as well | 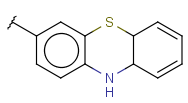 |
| Heterocyclyl(heterocycle)(since version 15.7.6) | - monocyclic or fused, aliphatic or aromatic ring with at least one hetero atom; carbon atom is also required | connection point at arbitrary position(s) | 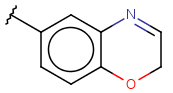 |
| Cyclyl (anycyclyl, anyring) | Any kind of ring regardless fuseness, aromaticity and hetero-carbo nature. | - | 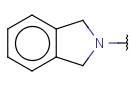 |
| Carbocyclyl (since version 20.20.0) | - monocyclic or fused, aliphatic or aromatic ring exclusively from carbon atoms | connection point at arbitrary position(s) | 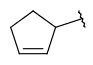 |
| RingSegment - | A part of a ring where every atom has only 2 ring connections. Non ring connections are allowed. The group does not represent a whole ring. | - | 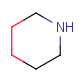 |
| HeteroSubstitutedAlkyl (HSA) | - at least one carbon atom- at least one hetero atom- single bonds- no ring bonds | connection point at arbitrary carbon atom(s) | 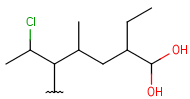 |
| Haloalkyl | - at least one carbon atom- at least one halogen atom- single bonds- no ring bonds | connection point at arbitrary carbon atom(s) | 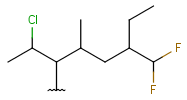 |
| Hydroxyalkyl | - at least one carbon atom- at least one terminal O atom- single bonds- no ring bonds | connection point at arbitrary carbon atom(s) | 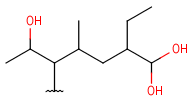 |
| Unknown group (UNK) | - | Any structure. Unknown structures are enumerated as the union of all other homology groups. | - |
| AnyAtom - | Any atom except hydrogen | - | C, N, O, P, S, ... |
| Metal (MX) | Any metal | - | U, K, Fe, Na, Ni, Al, ... |
| AlkaliMetal (AMX) | Alkali and alkaline earth metals | - | Na, K, Ca, Mg, ... |
| OtherMetal (A35) | Group IIIa-Va metals | - | Al, Ga, ... |
| TransitionMetal (TRM) | Transition metals excluding lanthanum | - | Fe, Ni, Zn, Co, Hg, W, ... |
| Lanthanide (LAN) | Lanthanides (including lanthanum) | - | Nd, Ce, Pr, ... |
| Actinide (ACT) | Actinides (including actinium) | - | U, Th, Pa, ... |
Subset rules between homology groups
-
alkyl, alkenyl and alkynyl are subsets of carbontree
-
carboalicyclyl, carboaryl, heteromonoaryl, heteromonoalicyclyl and fused hetero cyclyl are subsets of cyclyl
-
alkalimetal, transitionmetal, othermetal, lanthanides and actinides are subsets of metal.
-
all of the above groups are subsets of the "any" homology group
2. User-defined Homology groups
The homology groups are defined by the user, but there are some Predefined groups, too. User-defined Homology groups are represented by R-group definitions and during search these Pseudo atoms are translated to the corresponding R-group definitions.
These group definitions are customizable, the user can modify them or can make new definitions as well. Group names are treated as case insensitive, but in case sensitive file systems the definition files should be lowercase.
The following Predefined (User-defined) Homology groups are readily available in the system:
Halogen
Halogen elements: F, Cl, I and Br.
JChem's group name: halogen
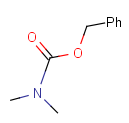
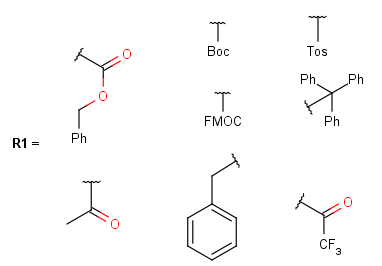
Protecting
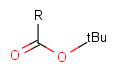
Protecting groups' definition file contains several definitions, each for protecting different functional groups. The protected functional group is defined by the neighborhood of the R-atom. When the R-atom has the same neighborhood as the "protecting" pseudo atom, then the group is replaced by the R-atom.
The conversion processes the group definitions in their order in the file. This means that more specific environments should be placed earlier. For example, a carboxyl protecting group definition should precede an alcohol definition, otherwise the alcohol definitions will be applied instead. Currently they are located in the following order:
-
amino
-
carboxyl
-
alcohol
Currently the system can't handle protecting groups having more than one attachment point, or groups where the heavy atoms of the functional group should be changed by the substitution. The readily available definitions contain amine, carboxyl and hydroxyl protecting groups.
JChem's group name: protecting
alias name: PRT
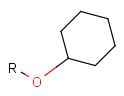
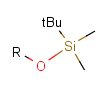
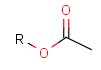
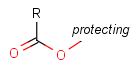
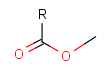
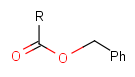
Some examples with different functional groups protected can be found on Table 2.
Table 2. Protecting group examples
| Protecting group | Represented examples | ||
|---|---|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
Any group
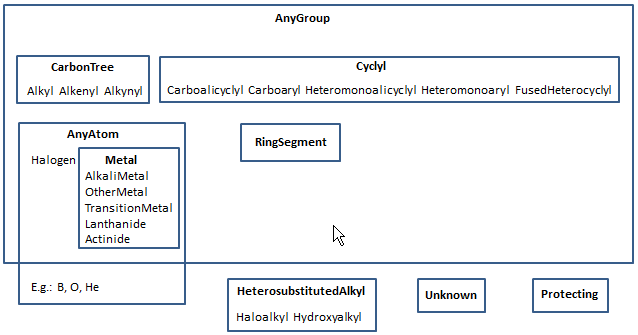
The union of all other homology groups except unknown and protecting. This union is represented by cyclyl, carbonTree, metal and halogen groups. If the group occurs in a ring then represents a ringSegment homology group.
JChem's group name: any
Search Options
Search options regulating the search behavior are also available:
Currently there is one regulating option: 'completeHG', which specifies if the part of the query side structure matching on the given group should represent an entire homology group or if substructures are also accepted. Of course in the incomplete case an entire structure can also match on the given homology group.
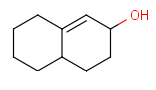
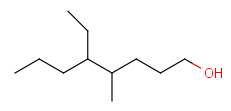
For example, if completeHG is set to true (default) an alkyl chain can't match on a cycloalkyl group, only a ring (system). The detailed behavior is found at the definition of the groups. And example is shown on Table 3.
Table 3. Complete and incomplete structures of Homology groups
| target | query | hit | |
|---|---|---|---|
| completeHG:y | completeHG:n | ||
 |
 |
 |
 |
 |
 |
 |
Markush Enumeration
To enable the enumeration of homology groups, the 'Homology Enumeration' option of Markush enumeration has to be switched on. Otherwise the 'Homology groups' are kept as 'Pseudo atoms'. This latter option might be useful for showing that these structures can't be fully enumerated.
Predefined Homology groups
For the Predefined groups, the R-group definitions specify the enumerable library as User-defined groups. I.e. these groups definitions can be customized. These structures are characteristic to the Homology group and encompass simple and large structures as well.
We have to emphasize, that these definitions are used only for enumeration and do not affect searching. As noted earlier, arbitrary structures fulfilling the requirements for the Homology group will match such a target.
Enumeration definitions contain two attachment points as default. After enumeration these are the atoms which connect to the first two neighbors of the group. If the enumerated Homology group's Pseudo atom has more than two connections, then further attachment points are added. These are put on atoms that have free valence and comply the requirements for externally connecting atoms of the given group. E.g. for 'aryl' only aromatic ring atoms can be the connection points. The atoms of the definition are investigated in the order of the Atom Numbers. If a definition does not have the sufficient number of such atoms, then it is rejected. When every definition of the homology group is rejected, an exception is thrown showing that the given homology group does not have any valid enumeration definition.
Customizing the (User-defined) Homology groups
Enumeration of User-defined Homology groups use the same ( customizable) R-group definitions as searching. User-defined Homology groups should have the same number of connections as in the definitions.
Customization modes:
-
To define NEW User-defined homology (or Protecting) groups under different file names from the Predefined files in the com.chemaxon-enumeration.jar.
-
To MODIFY the Predefined User-defined homology (or protecting) groups being in the com.chemaxon-enumeration.jar.
Enumeration-only type User-defined groups can be defined by the same way.
Modifying the Predefined Homology group definitions
The lib directories of the installed MarvinBeans and JChem contain com.chemaxon-enumeration.jar file. The default homology groups are defined there as user-defined groups and as enumeration-only groups.
Modifying the Predefined 'Enumeration-only' groupsDirectory chemaxon/enumeration/homology/enumeration_only in the com.chemaxon-enumeration.jar file. These groups are represented by these definitions during enumeration only.Modifying the new User-defined groupsDirectory chemaxon/enumeration/homology/user_def_groups in the com.chemaxon-enumeration.jar file. These groups are represented by these definitions during search and enumeration as well.
Location of user-defined homology group definition files
The default location of chemaxon_home directory of the user on different platforms:
-
Windows: %USERPROFILE%\chemaxon\ (in other words ..\Users\<USERNAME>\chemaxon)
-
Unix/Linux: ~/.chemaxon/
Location of "User-defined" (for search and enumeration) user-defined homology group definition files: chemaxon_home/homology/user_def_groups/
Location of "Enumeration-only" user-defined homology group definition files: chemaxon_home/homology/enumeration_only/
Note: Create the above two directories if they do not exist.
1. Defining NEW User-defined Homology (or Protecting) groups
-
Draw the desired group definition in MarvinSketch and save as mrv; the name of the new group should be specified by the name of the file; the name of the file must be in lower case;
See example nucleobase.mrv below:
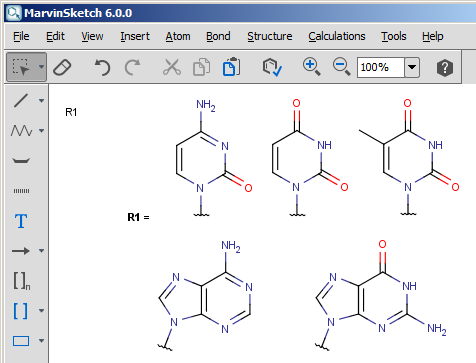
-
copy the mrv file into chemaxon_home/homology/user_def_groups/ .
The files of enumeration-only type User-defined groups should be placed into the directory chemaxon_home/homology/enumeration_only/ .
2. Modifying the Predefined (User-defined) Homology (or Protecting) groups
Modifying these files will affect searching/enumeration in case of User-defined groups and the enumeration in case of the Predefined groups.
The modified definition or the newly added group can also be dependent on the neighborhood (context-sensitive) as in the case of Protecting groups.
The modification of these definitions can be executed:
-
the same way as described above for the creation of the NEW User-defined Homology (or Protecting) groups, but the name of the mrv file must be the same as the built-in file name within com.chemaxon-enumeration.jar; copy the mrv file into chemaxon_home/homology/user_def_groups/
-
or by modifying the existing default file from com.chemaxon-enumeration.jar
-
Copy protecting group definition to the user's chemaxon library: e.g. from .../com.chemaxon-enumeration.jar/chemaxon/enumeration/homology/user_def_groups/protecting.mrv to chemaxon_home/homology/user_def_groups/
-
Open the newly copied file in the user's directory with MarvinSketch.
-
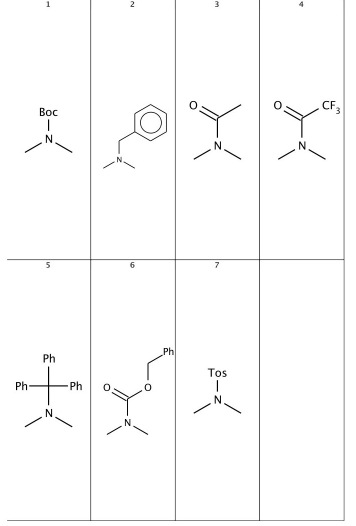
A dialog appears asking the index of molecule to open. Enter 1 because this contains the amino protecting group definition. If the proper molecule number is not known, all the definitions can be displayed using MarvinView.
-
Overwrite the structures, e.g. delete the FMOC group, see Table 4. The new definition will be used in searching and enumeration, see Table 4.
-
The files of enumeration-only type user-defined groups must be placed into the directory chemaxon_home /homology/enumeration_only/ .
If you would like to have different definitions for searching and enumeration of a user-defined group, then a separate file should be specified under the same file name in the " enumeration_only " dictionary as well. In this case the content of the " user_def_groups " will be used during searching and the content of the " enumeration_only " for enumeration.
If a definition is modified it comes into effect immediately, however the addition of a new group requires a restart of the Java Virtual Machine.
Table 4. Modifying amino protecting group definitions.
| overwriting the definition | sample markush file | enumerations |
|---|---|---|
 |
 |
 |
Properties of Homology groups
Some Homology groups have important properties. You might want to specify if the alkyl chain is branched, or any deuterium atoms are present. The Homology groups have a special property editing dialog where you can set the different properties. They include the followings (with the group to which it may be applied):
-
Deuterium and tritium count: for all Homology groups. The value should be given as e.g. D1-4T3, meaning the group contains up to 4 deuterium atoms and 3 tritium atoms.
-
Text notes: for all Homology groups.
Text format: Letters denoting different parameters followed by number ranges.
These entries are separated by commas (,). Specification of attachment atom type is also possible.
Possible parameters:
E
Number of double bonds
Y
Number of triple bonds
C
Number of carbon atoms
Hetero atom symbol
Number of occurrences of a particular heteroatom
X
Number of occurrences of heteroatoms not defined otherwise
Q
Number of occurrences of heteroatoms including defined ones as well (available from version 20.19.0)
HAL
Number of occurrences of halogen atoms (e.g., HAL1-5)
NR
Number of rings in a ring system
RA
Number of atoms in a ring system
atomic symbol
Presence of one attachment to the specified atom
atomic symbol
Presence of more than one attachment to the specified atom
Example: N1-3,NR4,E1-2,>>C
-
Branching: for chain Homology groups (BRA for branched, STR for straight chain).
-
Size: for chains. Chains are marked as low (C1-6. LO), mid (C7-10, MID) or high (C11-, HI) according to the length of the chain.
-
Saturation: for ring groups. They can be marked as saturated or unsaturated.
-
Ring type: for ring groups. They are marked as monocyclic (MON) or multicyclic (FU), or can be marked as 'not specified'.
Not specifying a property means that there is no restriction on that property.
Table 5. Available properties of homology groups.
| Homology groups | Size | Branching | D/T count | Ring type | Saturation | Additional Text Notes | |
|---|---|---|---|---|---|---|---|
| chain-like | Alkyl |  |
 |
 |
 |
 |
 |
| Alkenyl |  |
 |
 |
 |
 |
 |
|
| Alkynyl |  |
 |
 |
 |
 |
 |
|
| Haloalkyl |  |
 |
 |
 |
 |
 |
|
| HeterosubstitutedAlkyl |  |
 |
 |
 |
 |
 |
|
| Hydroxyalkyl |  |
 |
 |
 |
 |
 |
|
| CarbonTree |  |
 |
 |
 |
 |
 |
|
| cycle-like | Aryl |  |
 |
 |
 |
 |
 |
| Carboaryl |  |
 |
 |
 |
 |
 |
|
| Heteroaryl |  |
 |
 |
 |
 |
 |
|
| Carboalicyclyl |  |
 |
 |
 |
 |
 |
|
| Cyclyl |  |
 |
 |
 |
 |
 |
|
| Carbocyclyl |  |
 |
 |
 |
 |
 |
|
| FusedHetero |  |
 |
 |
 |
 |
 |
|
| Fusedheteroaryl |  |
 |
 |
 |
 |
 |
|
| Heteroalicyclyl |  |
 |
 |
 |
 |
 |
|
| Fusedheteroalicyclyl |  |
 |
 |
 |
 |
 |
|
| Heteromonoalicyclyl |  |
 |
 |
 |
 |
 |
|
| RingSegment |  |
 |
 |
 |
 |
 |
|
| anygroup-like | AnyGroup |  |
 |
 |
 |
 |
 |
| XX |  |
 |
 |
 |
 |
 |
|
| UnknownGroup |  |
 |
 |
 |
 |
 |
|
| atom-like | Halogen Metal AlkaliMetal TransitionMetal Actinide Lantanide AnyAtom |  |
 |
 |
 |
 |
 |