Extension Fields
Purpose of extension fields
Extension fields allow calculations to be performed on data from other fields. They can also access data from a datatree (child entity, grand child entity, etc) and from external sources like files, web services, databases, etc.
They allow to return chosen Java type, define custom query operators and custom icon. Java handler implementing ExtensionFieldHandler provides needed functionality and behaviour is adjusted using JSON configuration.
Extension field is an experimental feature, so expect API or behaviour changes.
How are calculations performed?
Provided ExtensionFieldHandler is called and returns all necessary information - field type, field dependencies, row values, custom operators and their evaluation, custom icon.
JSON configuration affects handler behaviour and can adjust field dependencies, constants, calculations, etc.
Where are calculations performed?
Calculation / data retrieval is done in IJC or in external data sources, depending on the handler implementation. Results are not stored in the database, but calculated dynamically as they're needed. Field values are cached.
How do I handle structures?
The structure field can be used as a dependent field. The data type is the
IJC Structure class.
The key thing you will probably want to do with it is to get the structure in either its original format (e.g. smiles, SDF, ...) or as
Marvin Molecule
object. Here are examples for each - both assume the Structure mol variable.
- As the original format:
mol.getEncodedMol()- Obtain the Molecule object and then call the
toFormat()method. You can do much more with the Molecule using the Marvin API:
mol.getNative().toFormat("smiles") // convert to smilesIn both cases a text value is generated (e.g. you would be defining the String.class type extension field). To display this as a structure you can use the structure renderer in the grid view or the molecule widget in the form view.
What about errors?
If an error occurs during field value retrieval or calculation, then it is thrown as usual.
Comparison with Calculated field
Main differences between calculated and extension fields.
| Feature | Calculated field | Extension field |
|---|---|---|
| Execution | Groovy script | Java handler |
| Storage | schema metadata | installed Java class |
| Data type | decimal, integer, text, boolean | any Java class |
| Error handling | returns null | throws exception |
| Search | available | available for basic types or using custom operator |
| Sort | available | n/a |
| Custom operators | n/a | available |
| Custom icon | n/a | available |
| External data sources | possible | easy to implement |
To add a new extension field:
A new extension field can be added by any of these methods:
-
In the Grid View
-
In Design mode of the Form View
Step 1. Install IJC plugin to make handler classes available:
Install IJC plugin containing Extension field handler classes.
For example download ijc-api-examples.zip and install ijc-api-examples\plugins\com-chemaxon-ijc-field-extension-demo.nbm.
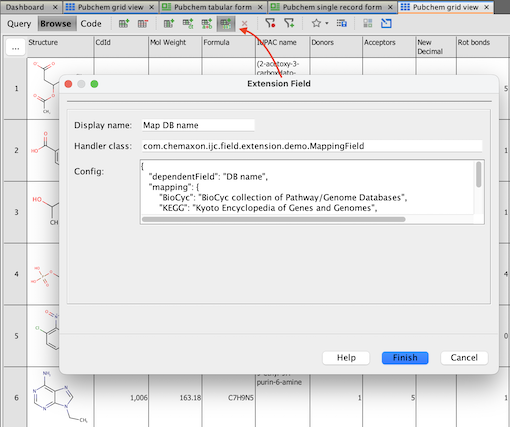
Step 2. Open new Extension Field dialog:
For example in a sample project, open Pubchem grid view and click the New Extension Field... (experimental) button.
Step 3. Fill-in values:
Enter field name, handler class and JSON configuration.
For example Map DB name, handler com.chemaxon.ijc.field.extension.demo.MappingField and configuration for mapping values from DB name field:
{
"dependentField": "DB name",
"mapping": {
"BioCyc": "BioCyc collection of Pathway/Genome Databases",
"KEGG": "Kyoto Encyclopedia of Genes and Genomes",
"MOLI": "Molecular Imaging Database",
"NIST Chemistry WebBook": "National Institute of Standards and Technology Chemistry WebBook"
},
"defaultValue": "undefined"
}Step 4. Create the Extension field:
Click Finish and check that a new field was created and values are filled-in.
For example values BioCyc collection of Pathway/Genome Databases are filled-in.
To edit an existing extension field:
Currently only a field name can be changed - either in the schema editor or in the widget customiser in the form or grid view.
Remove and add a new extension field if you want to change the handler and/or configuration.
Examples
1. Similarity field
Chemical similarity, a powerful tool in compound screening, is easily accessible in IJC via extension field.
-
Handler class to use for similarity is
com.im.df.impl.db.field.extension.use.SimilarityCalculator, config is to be left empty.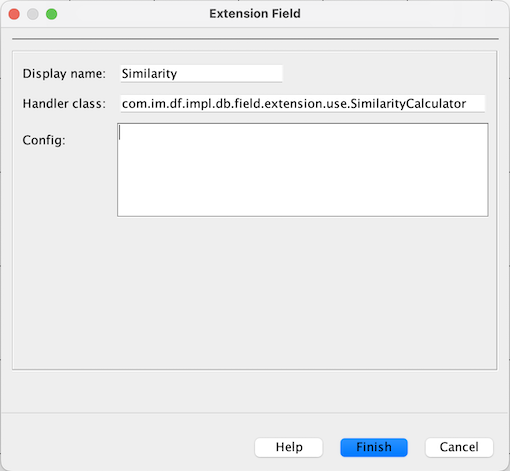
-
Newly added Similarity field will be empty since no query has run on it yet. In order to use Similarity search, switch to the Query mode and draw the molecule which you want to use as a standard for your query.
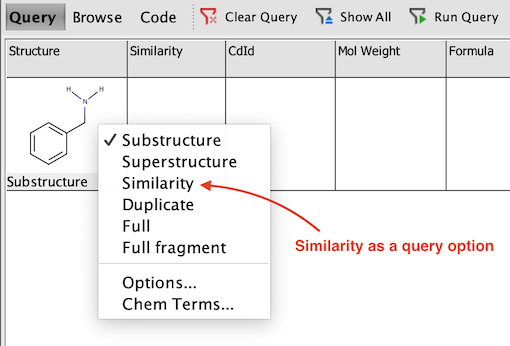
-
You will notice that default similarity is set to 0.5. In order to change the setting, right click the structure in Query mode and choose Options... Here you can modify Similarity threshold or change the method used to calculate Similarity (Screening config).
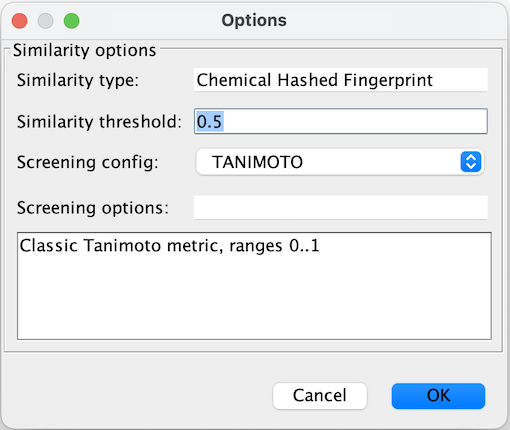
-
Once the query is run, compounds from your dataset with Similarity higher or equal to the threshold are shown. Screenshot below is an example of a query set in Pubchem demo on benzylamine with similarity threshold set to 0.4.
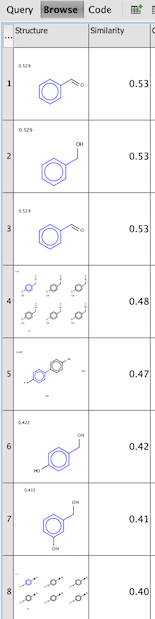
2. Chemical terms using extension field
Instant JChem offers an extensive array of chemical terms. You can conveniently integrate most of these desired chemical terms using the Chemical Terms field. However, this field comes with constraints: it only supports integer, text, boolean, and decimal value types. For calculating Chemical terms that return other types, like molecule, array and others, Extension fields under the com.im.df.impl.db.field.extension.use package can be used. We'll walk through how to use these by the way of specific examples.
You can view the entire list of Chemical terms provided in IJC here.
Example 1: Stereoisomers
The Stereoisomers chemical term produces all potential stereoisomers for each molecule in your dataset. This term creates field containing molecules (a data type not supported by the standard Chemical terms field). To accommodate this, Extension field has to be utilized.
Start by selecting the New Extension Field icon and input the subsequent details:
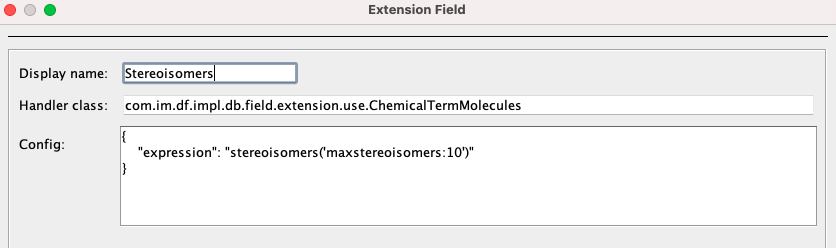
Package for these extension fields, com.im.df.impl.db.field.extension.use, remains constant for all chemical terms. However, the specific extension field handler varies based on the chemical term's return type. In this instance, since our return type is a set of molecules, the handler is ChemicalTermMolecules. See the Table below the Example 2 for more exemplary combinations of return type, handler and expression.
In the Config box, expression value can be written. The chemical terms function list also provides these expressions for each term. Following expression return at most 10 stereoisomers of a given molecule.
{
"expression": "stereoisomers('maxstereoisomers:10')"
}Upon clicking "Finish" new field containing stereoisomers is created.
Note: There is only one stereoisomer visible when the field is created. This can be adjusted via Widget Settings. Right-click the Stereoisomes field and select Customize Widget Settings. Here, desired number of rows and columns can be set.
In the following picture, two rows and three columns were set.
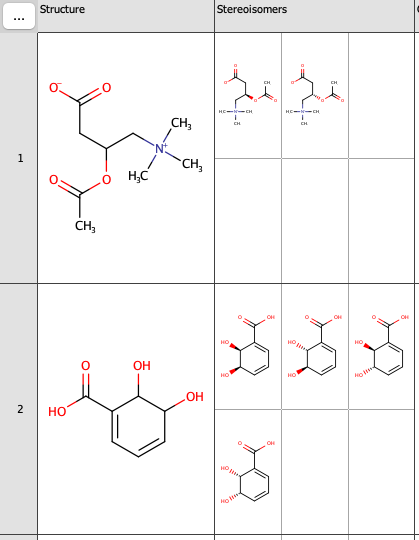
Example 2: Hybridization
Suppose you are keen to identify the hybridization on atoms within the molecules of your dataset. In this scenario, the created extension field will display a text array, with each row corresponding to an atom.
The handler class for this function is com.im.df.impl.db.field.extension.use.ChemicalTermArray.
The Config setup is:
{
"expression": "hybridization()"
}Inserting specific hybridization ('sp3', 'sp2', etc.) as an argument produces a field with atom indices having such hybridization.
{
"expression": "hybridization('sp3')"
}Result can be seen in the picture below.
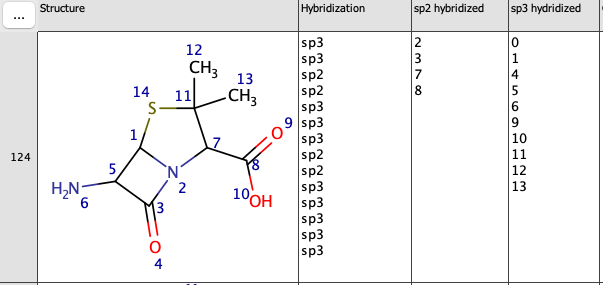
Please note, that MarvinSketch atom indexing (Structure field) starts from 1. However, with this expression field, atom indexing will commence from 0. In other words, the atom no. 1 in the Structure field corresponds to the atom no. 0 in Chemical term extension field.
| Chemical term return type | Extension field handler | Example expression |
|---|---|---|
| int[], double[] etc. | ChemicalTermArray | hbda('type:donorcount', 'pH:7.4') |
| Molecule | ChemicalTermMolecule | resonant(1) |
| Molecule[] | ChemicalTermMolecules | enumeration() |
| HergPlugin$HergResult | ChemicalTermHergBeta | hergBeta() |
| HergPlugin$HergResult | ChemicalTermHergClassBeta | hergClassBeta() |
3. Value mapping field
Example implementation of a value mapping can be found in ijc-api-examples.zip, downloaded from the IJC download page.
See MappingField.java in ijc-api-examples\sources\extensionField\Demo\src\com\chemaxon\ijc\field\extension\demo.
4. Mass Spectrum field
Advanced example is a Mass Spectrum field. Defines its own Java type for mass spectrum peaks, new icon, reads data either from a child entity or mass spectrum XML format, defines mass spectrum Similarity operator and provides the search. Its mass spectrum peaks can be rendered using a Canvas widget.
5. External data sources
Extension fields can retrieve data from any external data source. Detail data can be retrieved from a file or a different database. Chemical calculations can be computed using a web service.
6. Custom operators
Custom operators can bring new functionality - mass spectrum similarity search, biochemical subsequence search, etc. Evaluation happens in memory - values are first retrieved and then operator evaluated. Query returns matching results as returned by the operator.
Summary
These examples show that a wide range of functionality can be implemented using the extension field. Plugins can add custom functionality to IJC - extension fields retrieve data and canvas widgets render them.
We welcome feedback on this feature.