Upload compounds
Compounds can be uploaded on the Upload page in an SD file or you can paste as text.
 |
|---|
| Navigate with the arrows |
After the file to be registered has been selected, the user is allowed to navigate with the aid of arrows between the first records, while the accompanying data fields are displayed. The available data fields are determined based on the first ≈250 records. If more than 250 records are present, the [Scan more] button will be available. Each click on the [Scan more] button will process 250 more structures. If no more structures are available for examination, the button is not displayed.
{info} While the Scan more button is active the user is always notified about the number of compounds processed from the SDF
{info} Structure matching Compound Registration's structure matching logic is based on Chemaxon's JChem Stereochemistry. During structure registrations the JChem Global Stereo model is used.
Field mapping
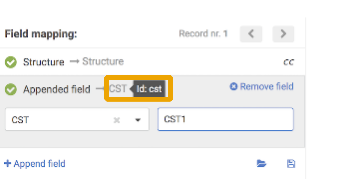
When uploading compounds, the data accompanying the structures can be e.g. LnbRef, Lot ID, PCN, CN, CST, Submitter, Salt ID, Salt multiplicity, etc. While the external ID fields (according to the actual business rules, e.g. LnbRef, Lot ID) can be configured to be mandatory or not for the registration, the structure (that can be an empty structure) is usually necessary to be able to initiate a registration (exception "No structures" in case of CSV files). Each column/field of the SD file can be mapped to any of the mandatory and optional fields of the structure object. The mapping can be set manually by selecting the appropriate parameter from the drop-down list above each column. E.g. "user" from the SD file can be mapped with "Lot submitter" from the DB table. The mapping can be saved in a text file using the [Save mapping] button and can be used again for other bulk upload registrations using the [Load mapping] button. When a field exists but contains a value just for some structures, there is a possibility to set a default value for the empty cases. After mapping the fields and setting a default value for the empty cases, the existing values will not be lost, just a value for the empty ones will be set.
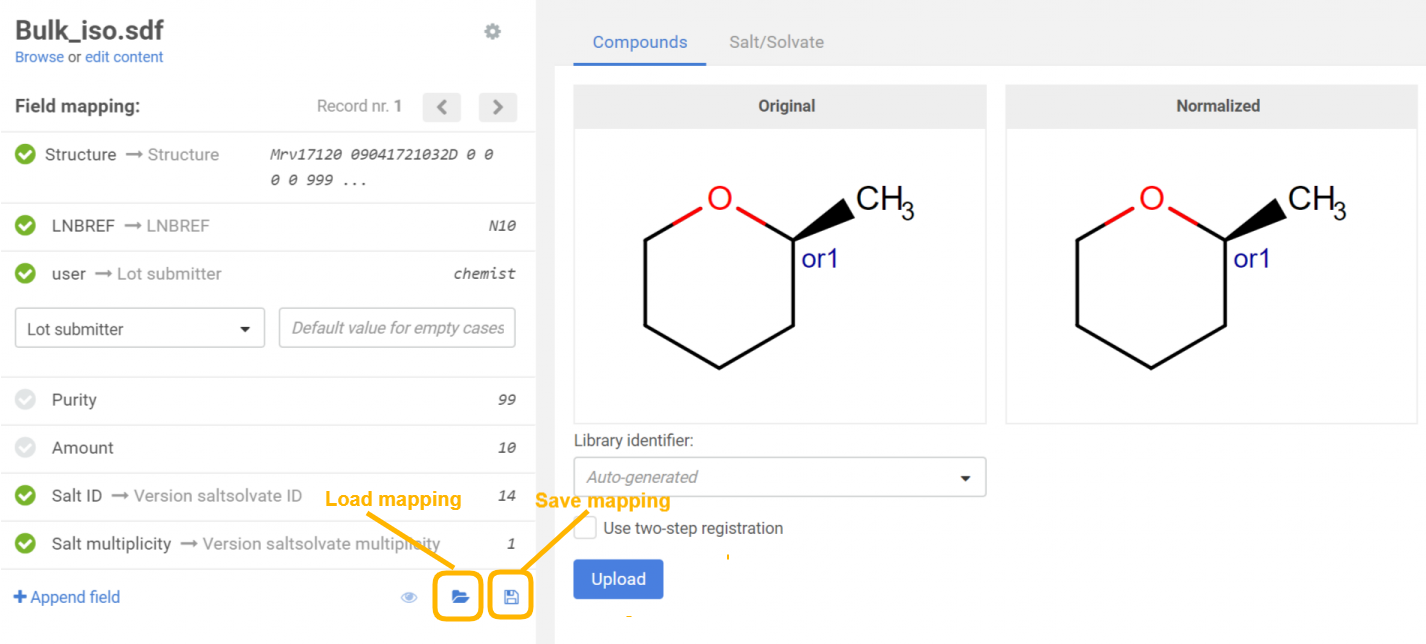 |
|---|
| Field mapping before uploading a file |
{info} Since version 20.8.0 during bulk upload you can only map fields that are configured for the source you want to upload with. Fields with validator: ‘required’ are also considered only if they are on the form of the given source. Configuration can be done on the Administration/Forms and Fields/Form Editor page.
{info} Since version 20.19.0 auto-mapping of fields during a bulk upload tries matching first to the field identifier of currently available fields, and if not found will perform a secondary matching to field display names. Field identifiers are displayed either next to field labels or as a tooltip when hovering over the respective field label in all relevant places on the bulk upload page.
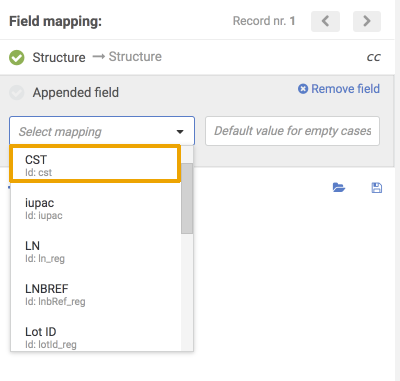 |
 |
|---|---|
| Select mapping drop-down with ids | Id tooltip |
Since version 21.3.0 for select fields the default value list is a drop-down, showing eligible entries for the given field.
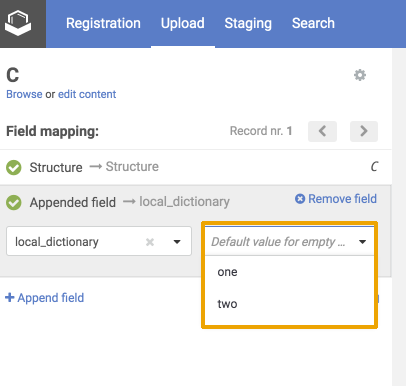 |
|---|
| Select field drop-down |
Uploading compounds with salt and solvates
- Uploading an SDF that contains compound and salt/solvate structures
Stripping salts/solvates from compounds is performed when the "Analyze Salt Solvate Fragments" Source dependent Registration option is ON. Only salts/solvates that are present in the Salts &Solvates Dictionary can be stripped.
- Uploading an SDF that contains compounds and salts Ids
During the upload, the user can provide the salt/solvate Ids and multiplicities for each compound structures if needed. Salts/solvates from the Salts &Solvates Dictionary can be referred by their Id and multiplicity in the SDF within a data field that is going to be mapped with the "Version salt/solvate ID" and "Version salt/solvate multiplicity" fields (Figure).
The result of the upload process is always summarized in the:
Uploading with specified Ids
During a bulkload process it is possible to set specific Ids (PCN and/or CN) for the registered compounds. For this, you just need to map the PCN and/or CN fields to the corresponding columns of the SDF file.
{info} In the case of SDF files the structure field is mandatory. When the "structure" part is "empty" a No structure registration is triggered.
{info} If the provided structure and the Id are not matching, the structure will be considered during the registration and the mapped Id is stored within an additional data field in the Uncategorized data section.
{primary} The Id field with the value from the Uncategorized Data section can be deleted after the registration.
Uploading no structure compounds
-
Uploading No structures under different parent compound Ids
No structure compounds can be sucessfully uploaded through an SDF if that contains empty structures and have the structure field mapped.
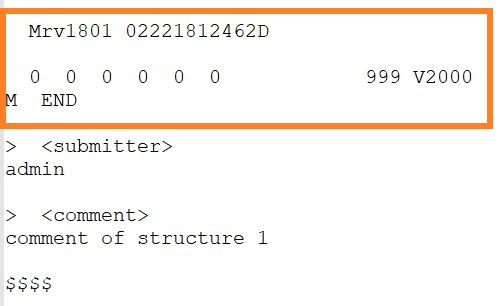
Example SDF that contains empty structure Each empty molfile will be registered as a new compound in the Registration system.
Similarly, in order to bulk register No structure compounds with CST, the value from the SDF should be mapped with the "CST" field in the application.
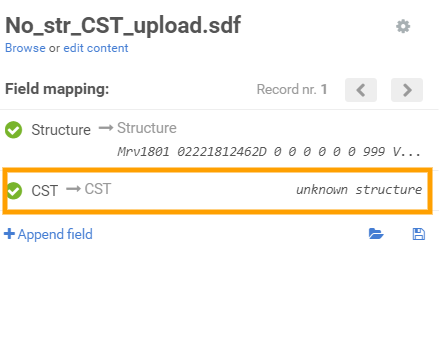
Mapping the CST on the Upload page -
Uploading No structures under the same parent compound Id (bulk register new lots)
No structure compounds can be sucessfully uploaded through an SDF under a specific parent compound Id, if the Id value is mapped to the "PCN" field in the system.
If the same CST is provided for a set of No structures within the SDF, the records will be registered under the same parent compound Id.
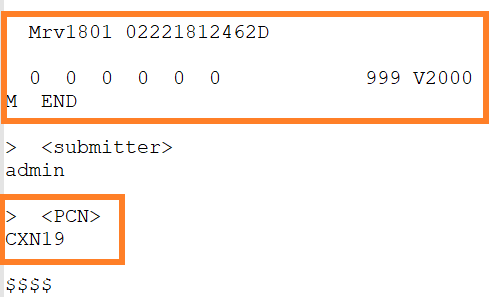
Example SDF that contains empty structure and mapped PCN
Uploading multi-component compounds
Please note, that the Bulk Upload does NOT support the registration of multi-component compounds. In the case when the loaded SDF file contains multiple components within one structure, this will be registered as a "single" compound consisting of more components (not as a Mixture, Formulation or Alternate). Or, as a solution, a multi-component checker can be introduced, in which case these records will not be autoregistered but will fall to the Staging area, from where these can be manually registered as multi-component compounds.
Uploading Invalid structures
Unreadable file format : When a molfile cannot be parsed and no information can be extracted from it (because it might be corrupted or does not follow the expected file format specification).
Inconsistent file format : Even though a valid molfile (can be parsed according to syntax specification, passes format validation) is present, it can happen that the molecule described by the file does not make any sense and is likely wrong.
Unsupported file format : Chemaxon may not know that a section of the file contains structure-relevant information and simply skip parsing that information. Often caused by modifications to the file format specifications introduced by vendors or organizations, that are not widely used and not sufficiently described publicly. In other words: we might miss extracting data, that is not supposed to exist according to the publicly available file format specs and we cannot assume what type of data and what its context might be.
Using Compound Registration invalid structures can be uploaded successfully. The compounds will be stored as "No structures" while the original structure as the text will be stored as additional data. Later the "No structures" can be amended on the parent compound level and the correct structure can be provided (the Ids will be kept).
In order to prepare the system for storing the original (invalid) molfile as a text, an additional data field should be created (Administration/Forms and Fields) that has the "errorStructure" as field identifier.
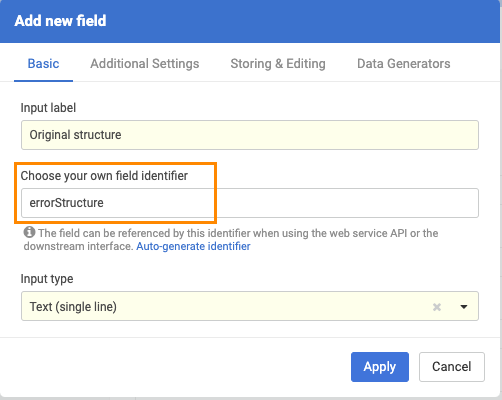 |
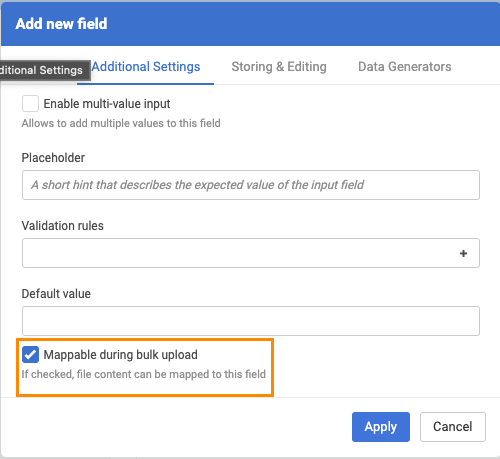 |
|---|---|
| New field configuration |
On the Upload page the previously created additional data field can be appended:
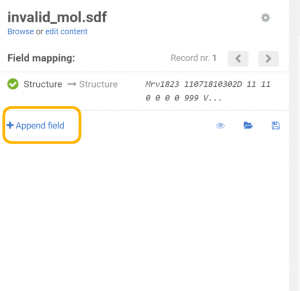 |
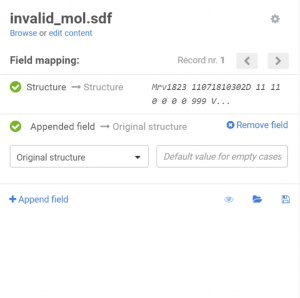 |
|---|---|
| Appending an additional field |
After uploading the file, the records that have invalid structures will be converted to "No structures" and will have the structure as text in a field stored as additional data to the compound.
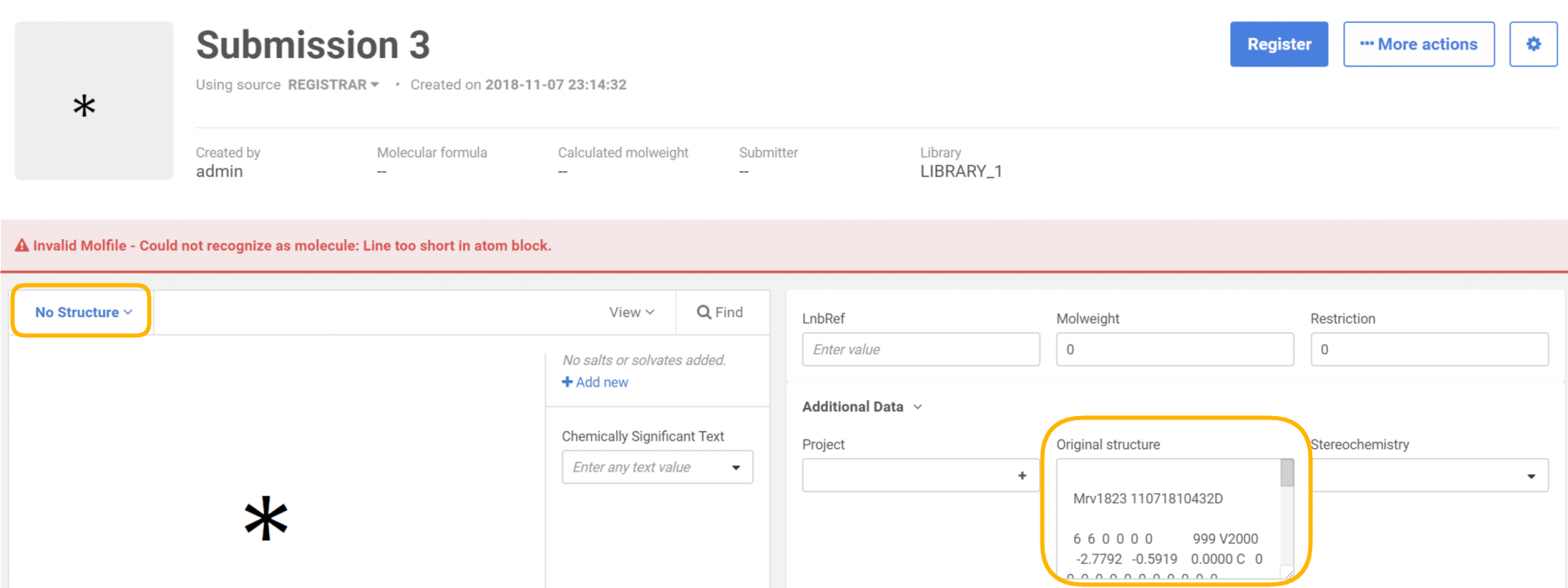 |
|---|
| During registration on the Staging area/Submission page |
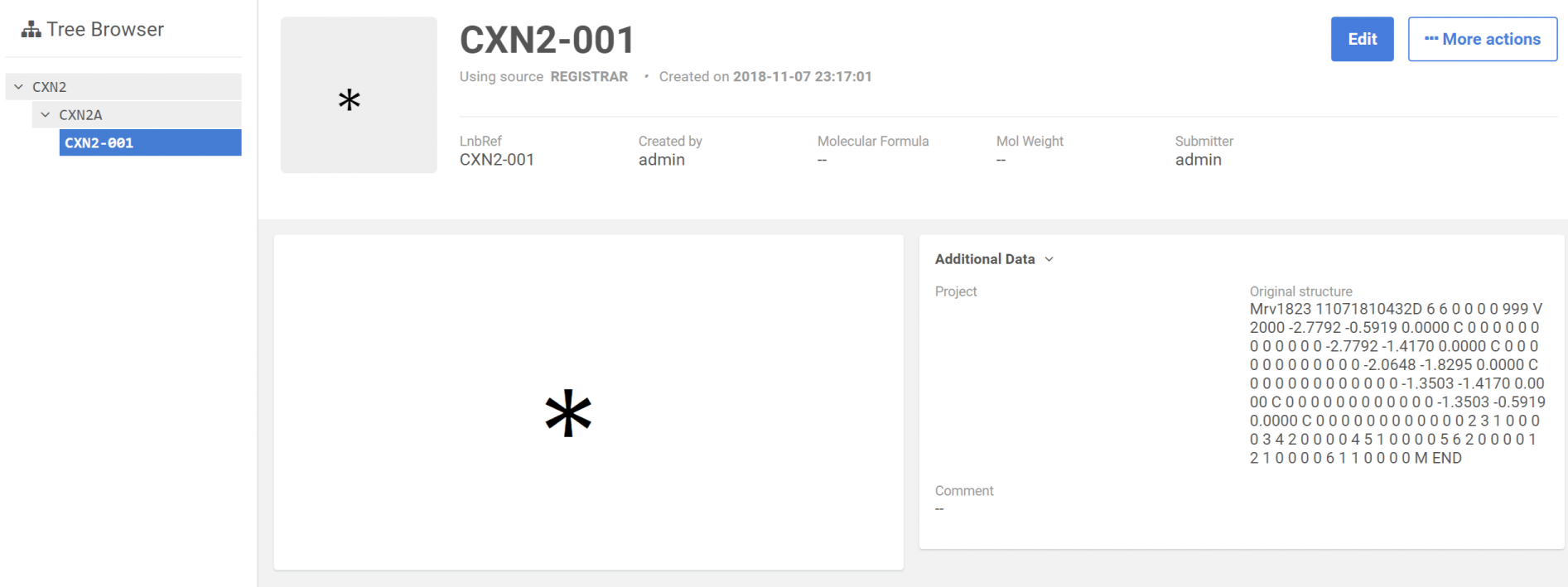 |
|---|
| After registration: Browse page |
The result of the upload process is always summarized:
Upload configuration
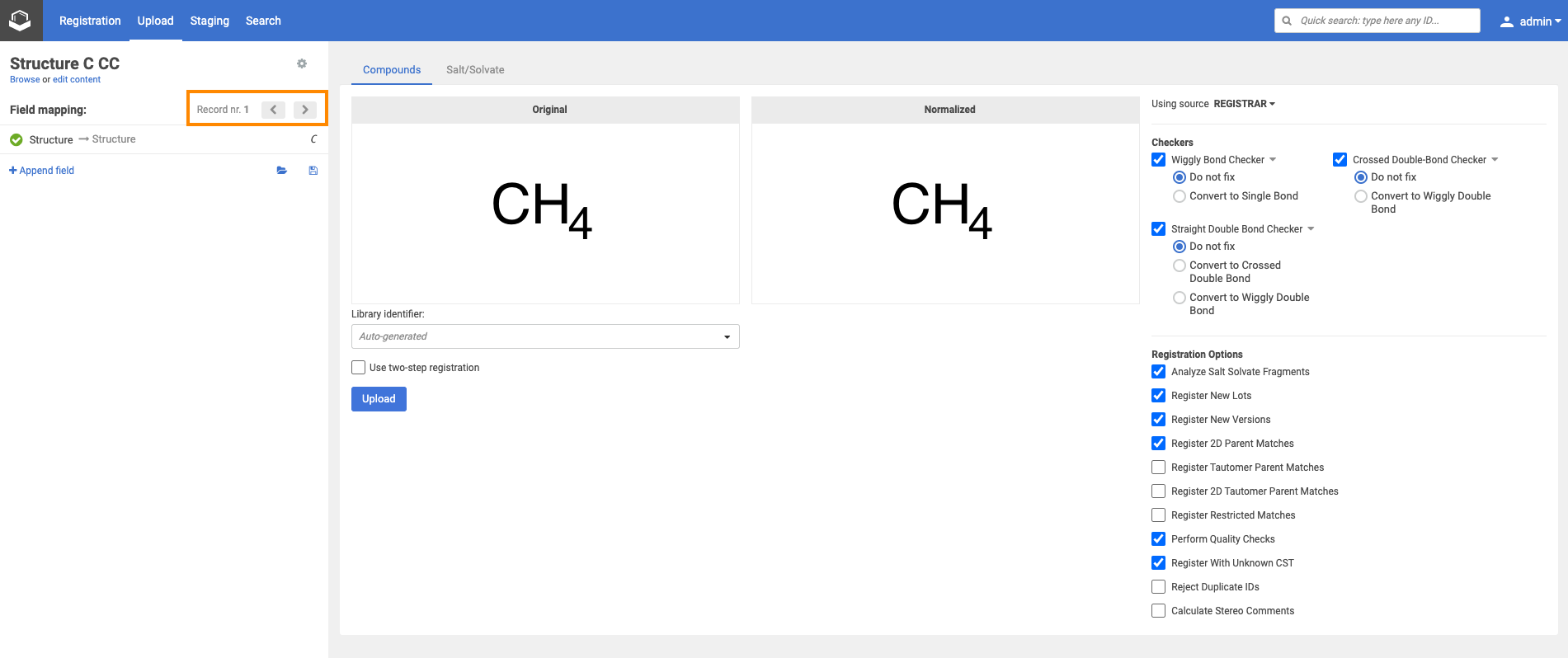
For a bulk upload process, the source can be set. According to the selected source, Checkers (and fixers) and Registration options become available, that will be applied for all the structures of the selected SD file.
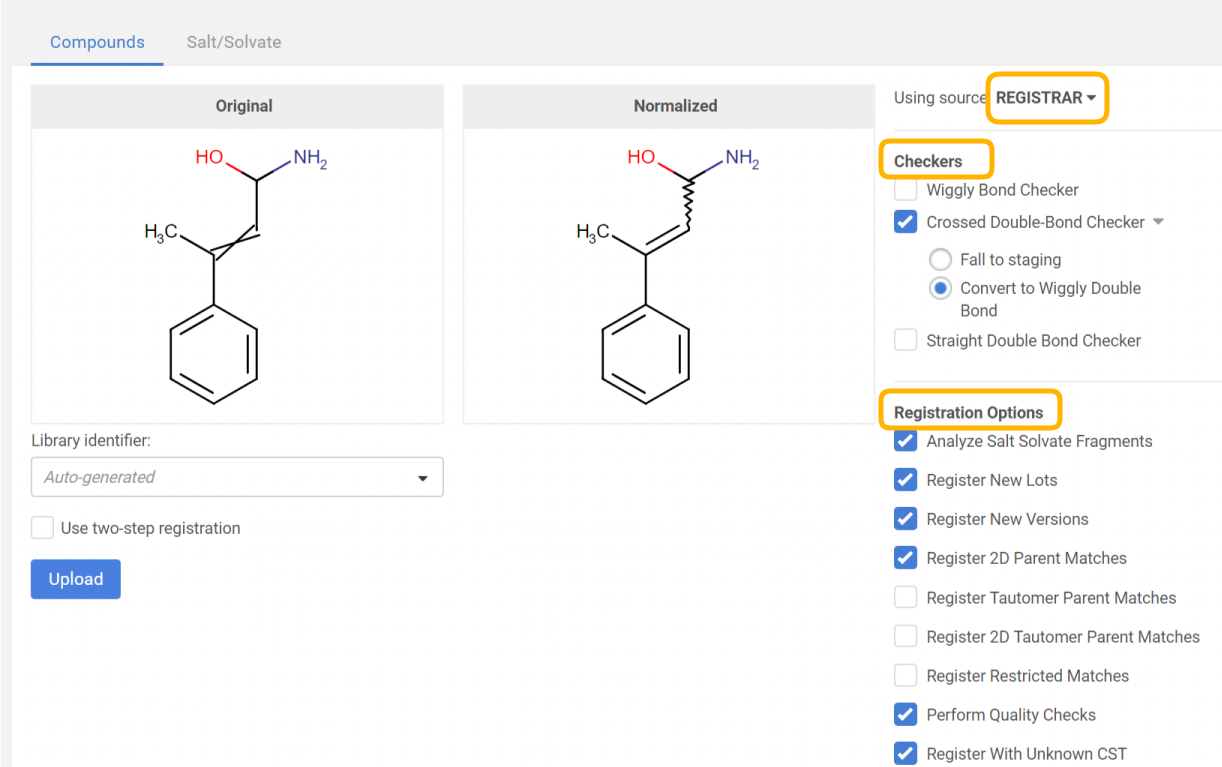 |
|---|
| Applying a structure checker and fixer. The original and normalized structures are displayed. |
By clicking on the [Upload] button, the registration process will run according to the autoregistration process, with the only difference, that here the selected structure fixer options will be applied to the structure before the actual registration and the "normalized" strructure can be compared with the "original" one.
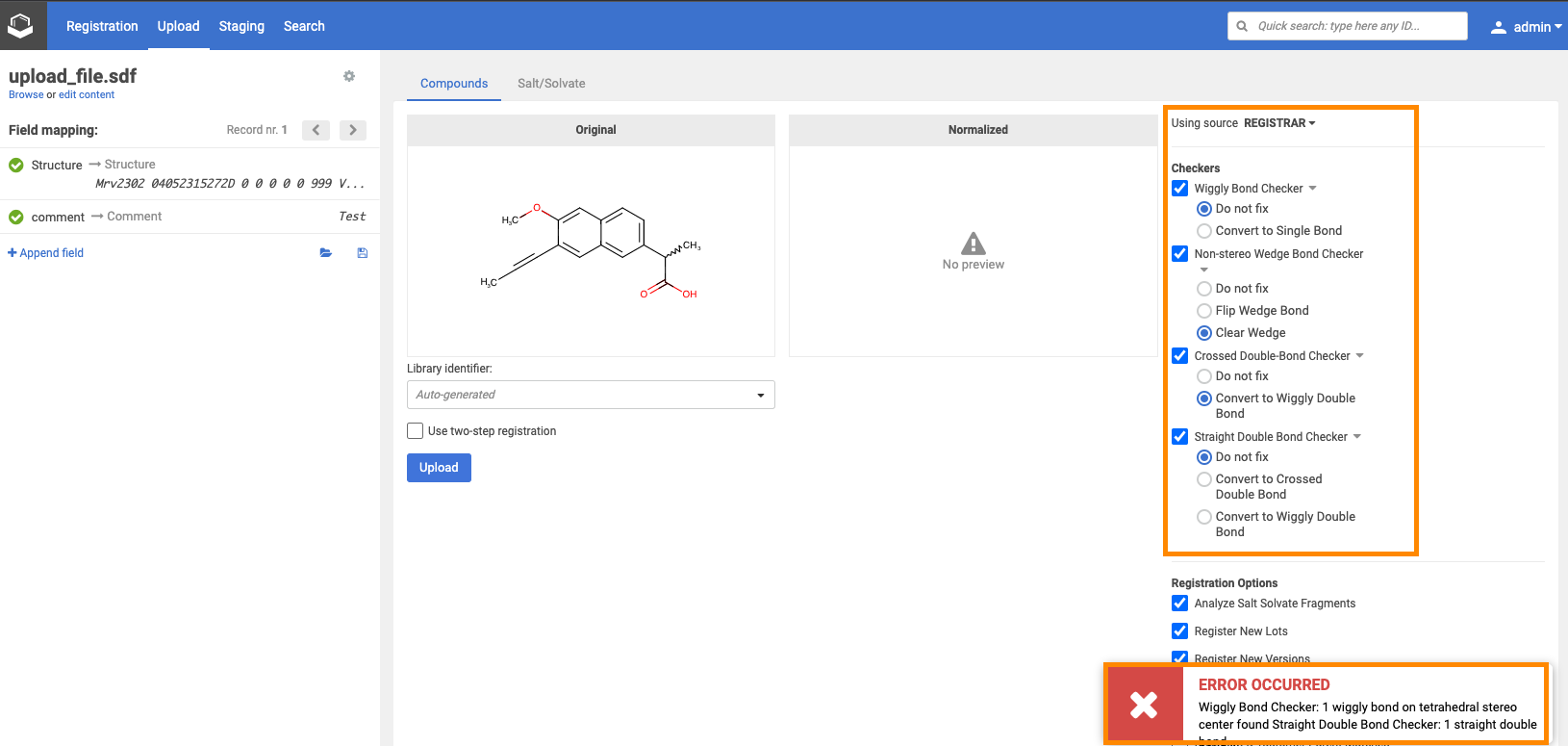
Source-based checkers
{info} Before version 22.11.0: Similar to the advanced registration process source-based checkers and fixers are not applied by default during the bulk registration, but need to be enabled manually.The Quality Checks defined at the system level will always run.
{info} Since version 22.11.0: All source-based checkers are enabled by default in advanced mode registration, bulk upload, and submission pages. Please find more information below.
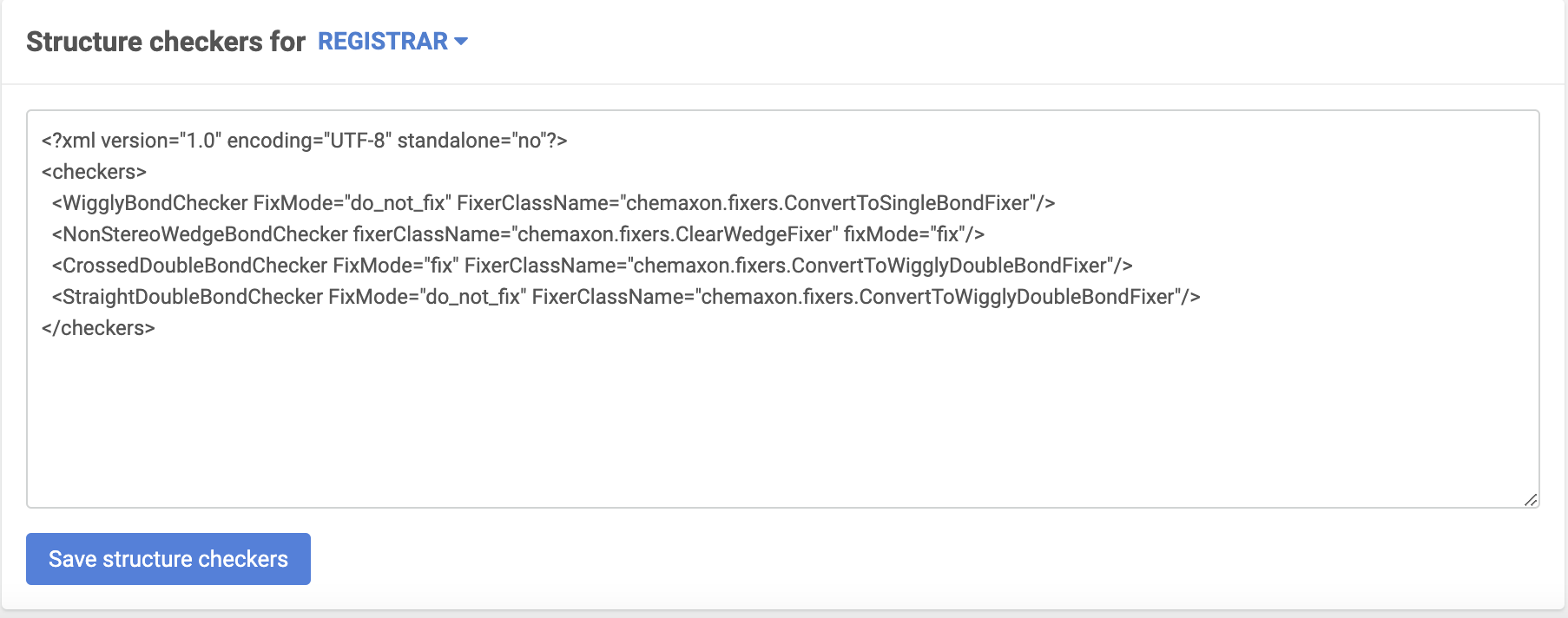 |
|---|
| Source-based checker configuration |
 |
|---|
| Source-based checkers on the Upload page |
All source-based checkers are enabled by default on the Upload page.
- If fixMode is set to "fix", the fixer provided in configuration is selected as default.
- If no fixerClassName is given, the "Do not fix" is selected as default
- If set fixMode is valid and different from "fix", e.g. "do_not_fix" or "ask", "Do not fix" is selected as default.
Default source-based checker setting can be modified manually on the Upload page.
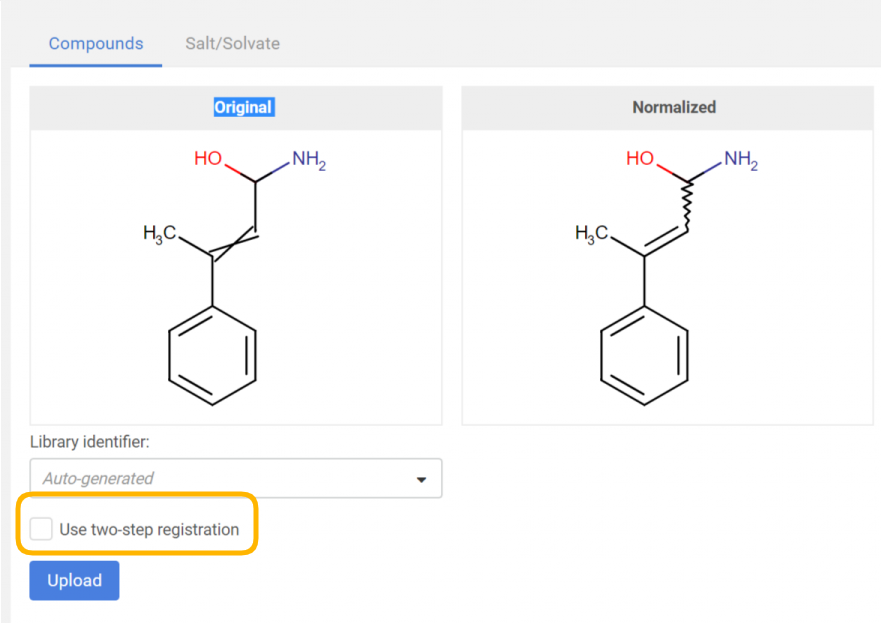
Two-step registration
The two-step registration option allows the user to pre-register the compounds to the staging area in order to check them before registration.
 |
|---|
| Two-step registration option on the Upload page |
Please find more details on the Upload summary page - Ready for registration and Failed submissions part.