ChemCurator User Guide
- ChemCurator Application
- Main Menu and Toolbar
- Panels and Views
- Create new project
- Chemical data extraction
- Fixing annotation errors
- Share and Export functions
ChemCurator Application
![]() ChemCurator is a desktop application of Chemaxon for computer-aided chemical information extraction. ChemCurator is a standalone desktop application. For running this application, you need to download and run ChemCurator installer. Short video tutorials demonstrating the main functionality are available here.
ChemCurator is a desktop application of Chemaxon for computer-aided chemical information extraction. ChemCurator is a standalone desktop application. For running this application, you need to download and run ChemCurator installer. Short video tutorials demonstrating the main functionality are available here.
Main Menu and Toolbar
The main menu contains "File", "View", "Window" and "Help" elements.
-
File
-
New Project...
 (Ctrl+Shift+N)
(Ctrl+Shift+N) -
Open Project...
 (Ctrl+Shift+O)
(Ctrl+Shift+O) -
Open Recent Project
-
Close Project
-
Import Project from ZIP...
-
Export Project to ZIP...
-
Restart
-
Exit
-
-
Edit
-
Undo
-
Redo

-
-
View
-
Link Project View
-
Show Only Editor (Ctrl+Shift+Enter)
-
Full Screen (Alt+Shift+Enter)
-
-
Tools
-
Plugins...
-
Options...

-
-
Windows
-
Projects
-
Checker View
-
Reset Windows
-
Close Window
-
Close All Documents
-
Close Other Documents
-
Documents...
-
-
Help
Panels and Views
Most of the panels and views in ChemCurator are optionally resizable, or can be moved to different location or to a different screen depending on your preferences. The default settings can be restored by the Reset Windows function.
Project explorer panel
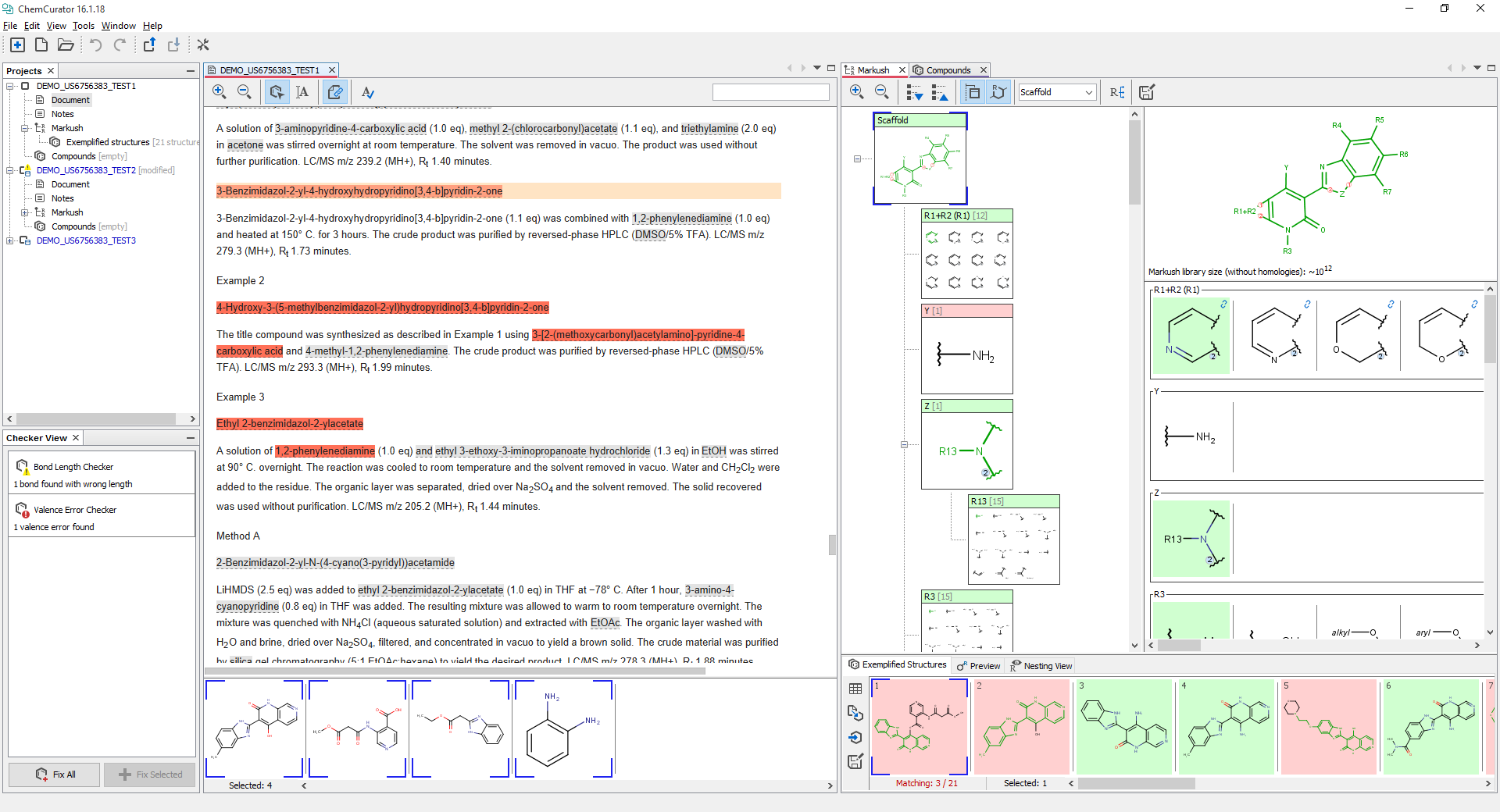
Project explorer panel displays the opened projects and represents the project's structure in a tree-like hierarchical way. Every project contains a single document, but you may add as many Markush structures and compound lists as you want. All Markush structures automatically obtain an Exemplified structures list.
Document view
Document view is the viewer component of the annotated documents and the related selections. The recognized chemical entities are highlighted by gray. In structure selection mode ![]() users can select recognized chemical structures by clicking on any highlighted component or select a larger part of the document by pressing left mouse button and dragging it over the targeted part of the document. The selected structures are highlighted by red and displayed under the document in the selection panel. In text selection mode
users can select recognized chemical structures by clicking on any highlighted component or select a larger part of the document by pressing left mouse button and dragging it over the targeted part of the document. The selected structures are highlighted by red and displayed under the document in the selection panel. In text selection mode ![]() users can select the document text directly. Document linking
users can select the document text directly. Document linking ![]() turns on the automatic scrolling of the document based on the structure selections in the editor views. With
turns on the automatic scrolling of the document based on the structure selections in the editor views. With ![]() and
and ![]() document's zoom level can be changed.
document's zoom level can be changed.
Compounds view
Compounds view is the display component of compounds lists. It can handle chemical structures and additional columns storing related information. Data can be edited by double-clicking on any of the cells.
Markush Editor view
Markush Editor View is the display component of the Markush structures and related exemplified structures. Markush Editor View is based on the same component as the standalone desktop application called Markush Editor. Therefore, the details of editing Markush structures are available in Markush Editor documentation. An additional bottom line of this view contains the exemplified structures related the Markush structure. Exemplified structures can be continuously validated against the Markush structure (this behavior is the default option, but it can be disabled for performance reasons). The exemplified structures that match the Markush are highlighted with green background, while non-matching structures are highlighted in red.
Structure checker panel
Structure checker panel displays the structure drawing errors and warnings related to the active editor component. In the case of an error, an exclamation mark appears in a red circle, and in the case of warnings, a yellow triangle appears. By clicking on the checker items you are able to choose between the available automatic fixer options. You are able to fix the issues one-by-one with the Fix Selected button or all together with the Fix All button.
Create new project
In ChemCurator, every project represents a single document and the extracted chemical information that belongs to this document. ChemCurator offers multiple project creation options based on different search formats. Independently of the original format, all documents are converted to an annotated HTML document preserving the structure and layout of the original source. The time required by the annotation process strongly depends on the format, size, and content of the original document. The new project wizard is available from File>New Project... or from the main toolbar with the  icon.
icon.
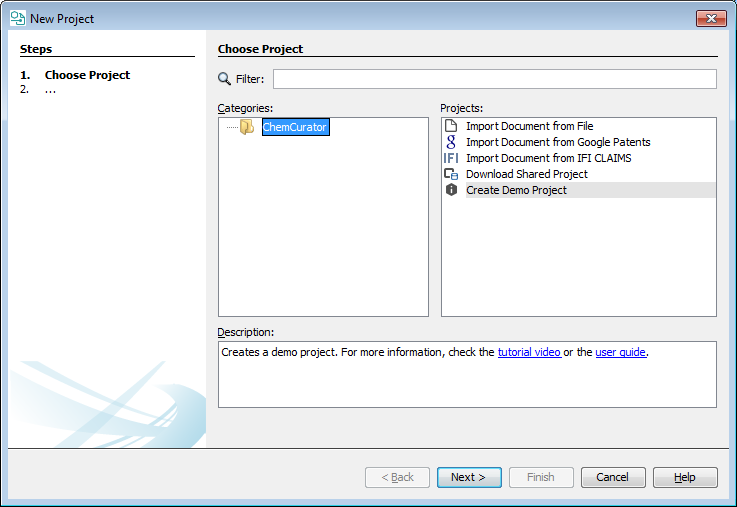
Import document from file
The project can be created from a file stored in your local machine. ChemCurator can process PDF, HTML, XML and TXT documents.
Import document from Google Patents
Patent documents can be imported directly from Google Patents by using the publication number of the document. The import wizard automatically tries to find the corresponding document in Google Patents and automatically downloads the HTML version of the patent. For most of the non-English patents, machine-translated English versions are available via Google Patents. If you want to download the original version, select Original from the language preferences.
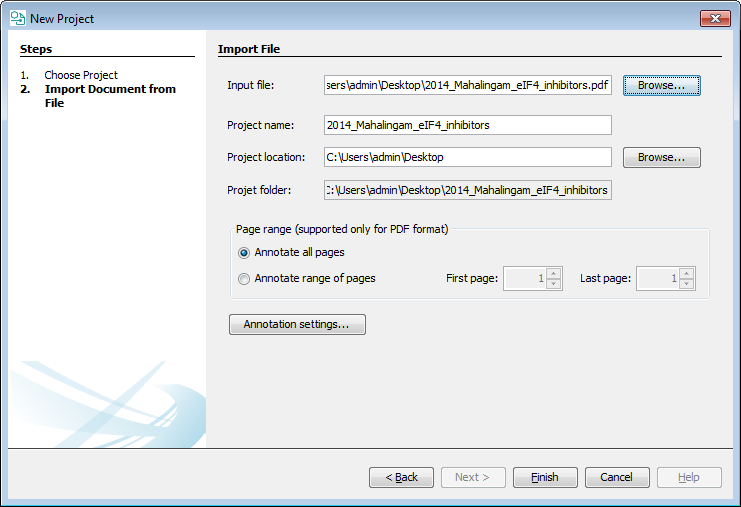
Import document from IFI Claims
If you have IFI Claim access, you can also import documents directly fromIFI Claims. The import wizard automatically tries to find the corresponding document in IFI Claims and automatically downloads the HTML version of the patent.
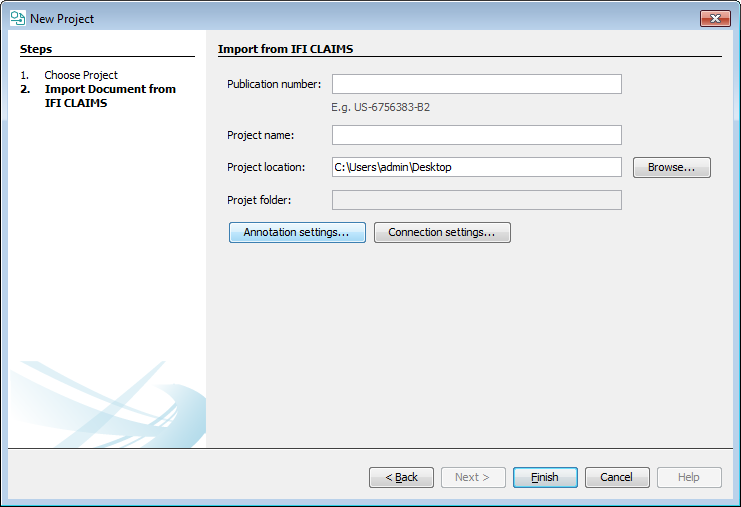
Create demo project
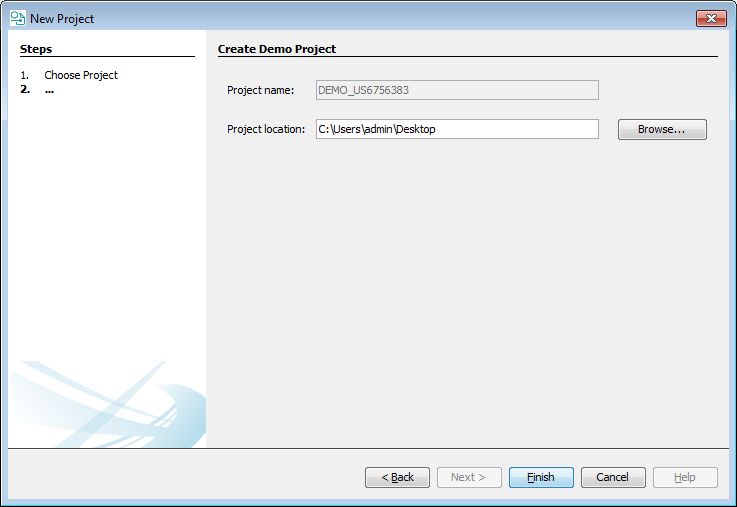
With this function, an example project can be created containing the annotated version of US6756383B2 patent document from Google Patents and some curated data including a Markush structure and compound list.
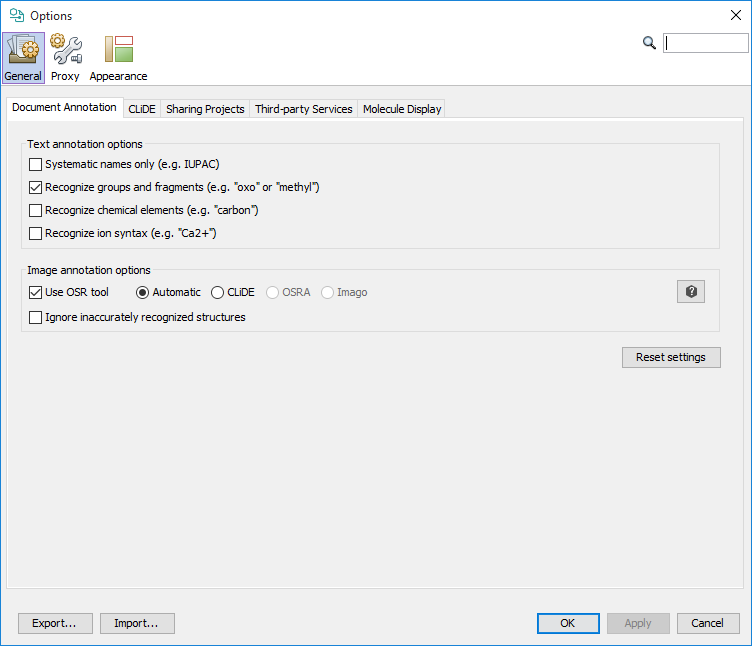
Annotation configuration
With annotation configuration, you are able to fine-tune the annotation parameters according to your needs. The settings panel is available from File>Options... or from the main toolbar with the ![]() icon.
icon.
Chemical data extraction
ChemCurator offers multiple functions to help in the recognition and extraction of the relevant chemical information from documents.
Create new Markush or Compounds list
ChemCurator supports two types of chemical information: Markush structures, and Compound lists. Markush structures are always created together with a linked special compound list that stores the corresponding exemplified structures.
Manual structure extraction
Any annotated structure can be selected from the document. After selection, it can be moved using drag-and-drop from the selected structures view to editor components.
Compounds extraction wizard
Compounds extraction wizard is available in Compounds and Markush view. This wizard can help to automatically find and extract a large number of chemical structures from the documents. In the first panel of the wizard, some basic filter criteria are available.
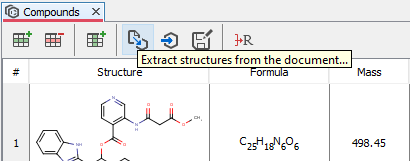
The extraction process can be parametrized with some filter options.
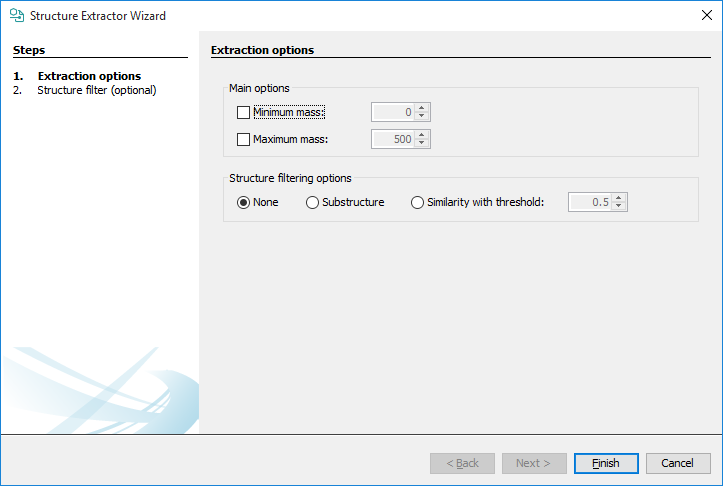
Main options:
Minimum mass : set the minimum molar mass for the structures to be extracted.
Maximum mass : set a maximum molar mass for the structures to be extracted.
Structure filtering options:
None : structure filter is ignored.
Substructure : a substructure filter criteria can be set after clicking on the Next button.
Similarity with threshold : a similarity filter criteria can be set after clicking on the Next button. MCS-based similarity calculation is executed in the background and the structures are filtered according to their Tanimoto similarity compared to the molecule that is drawn after clicking on the Next button.
If Substructure or Similarity with threshold is selected, then clicking on the Next button navigates to the second tab of the extraction wizard. In a case of Similarity with threshold , only exact compounds can be used as a filter without any variability. In a case of Substructure filter, however, atom lists, bond lists, and any query properties can also be used.
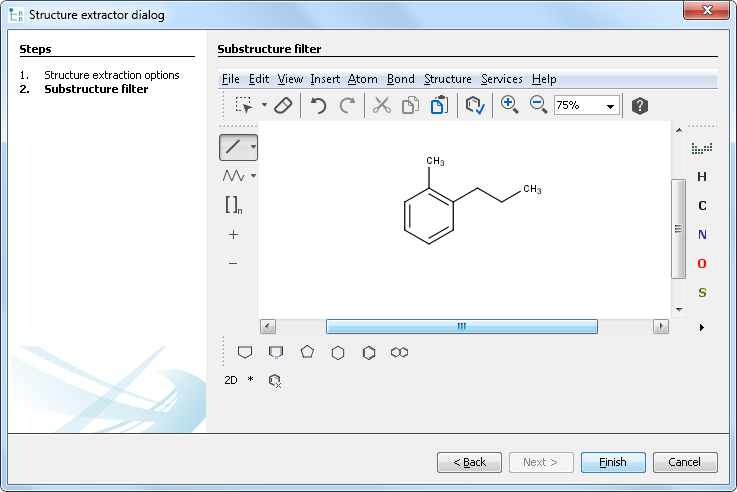
After clicking on the Finish button, extraction is started. In the case of Similarity with threshold , an additional column is also added to the extracted compound list containing the similarity values.
Additional data extraction
Compounds view is capable of handling not only the chemical structures, but also the related assay data, properties, comments, etc. You can manually add this information to the compounds lists using the Create new column function.
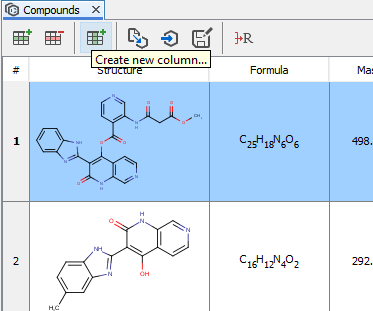
A simple dialog opens where the name and type of the new column can be selected. The newly created column can be edited by simply double clicking on it.
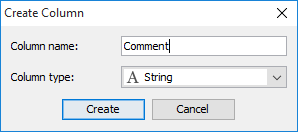
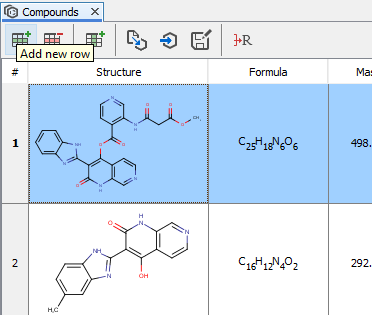
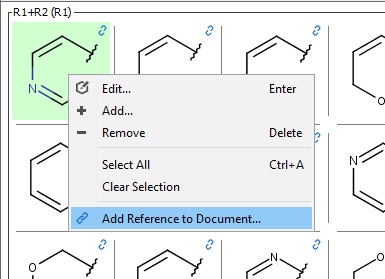
Add structures manually
Markush fragments and compounds can be added manually from fragment and compound list's context menu and with the Add new row menu item of the compounds view.
 |
 |
 |
|---|
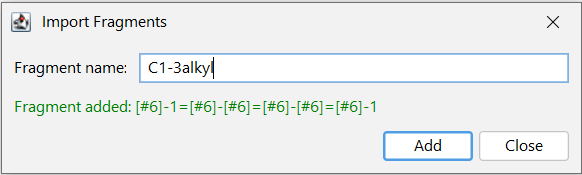
Fragment definitions can also be entered based on their name. In the right-click context menu the Import Fragments by Name... function allows to enter a chemical name, a homology name, SMILES or SMARTS (e.g. benzene or CC). The function can be reached with the Ctrl + I shortcut. Properties of homology groups are also recognized. It is also possible to consecutively add several different fragment definitions from one window as shown on the screenshot.
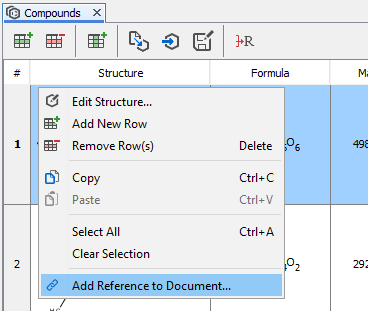
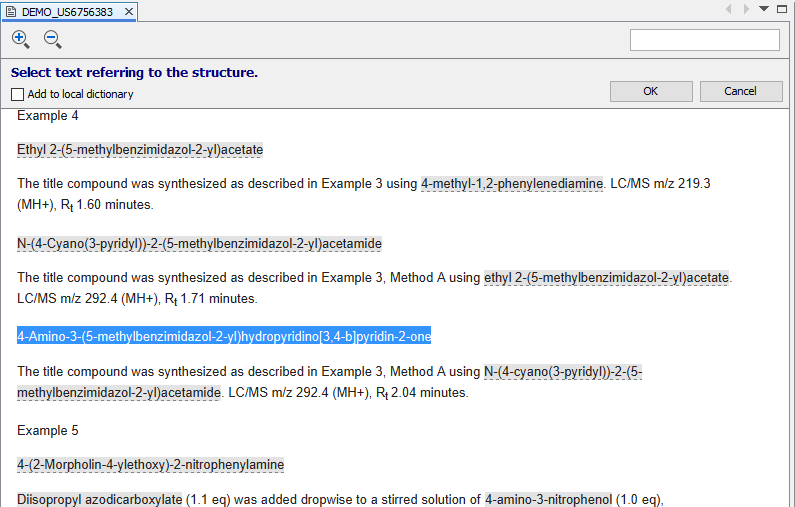
Manually added compounds can be linked to the corresponding part of the document. After a right click on any structure, you can select Add reference to document... function to specify the corresponding part of the document. After starting this action, document view enters reverse linking mode and any part of the document can be selected. After selecting the corresponding part of the document and clicking on OK, the selected part of the text will be marked as a chemical entity and linked to the manually added compound. If Add to local dictionary check box is also checked, the selected text and the linked compound are added to ChemCurator dictionary and will be recognized next time during annotations.
Fixing annotation errors
The accuracy of structure recognization is not 100%, so annotated documents almost always contain some unrecognized structures or not perfectly recognized structures.
Fixing misrecognized structures
Text-based and image-based structures that are not recognized correctly can be fixed by selecting the problematic structure in the document view, simply double clicking on it on the selection view or right-clicking on it and choosing the edit option.
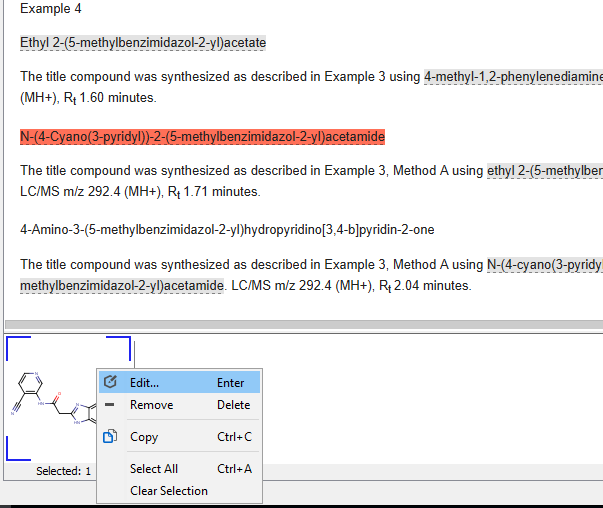
Fixing unrecognized structures
Unrecognized structures can be annotated by the Fix annotation menu item.
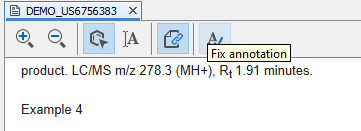
By clicking on this button, the document view enters reverse linking mode and any part of the document can be selected.
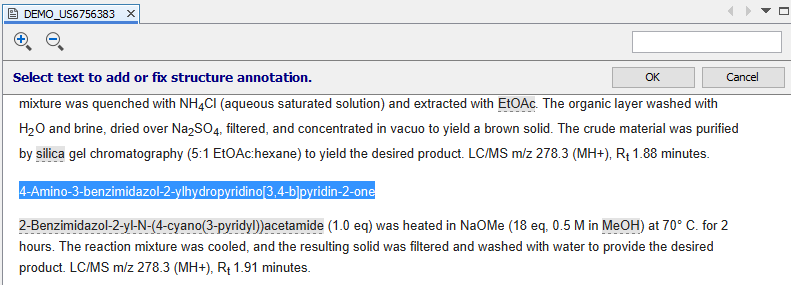
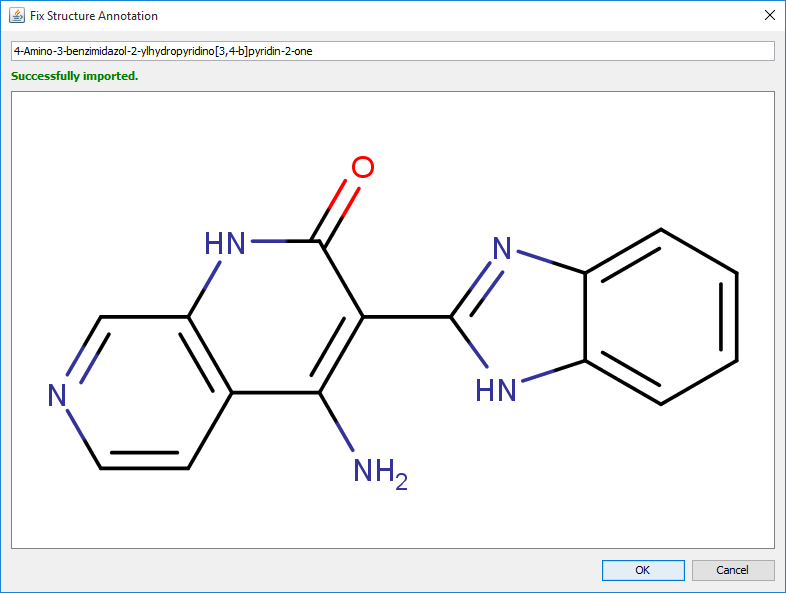
After clicking on the OK button, an interactive fixing dialog opens. If the modified text can be recognized, the recognized structure appears under the text input field. The structure immediately follows any modification of the text. Potentially problematic parts of the chemical names are underlined. After successful fixing, the recognized chemical structure can be added to the corresponding part of the document by clicking on OK button.
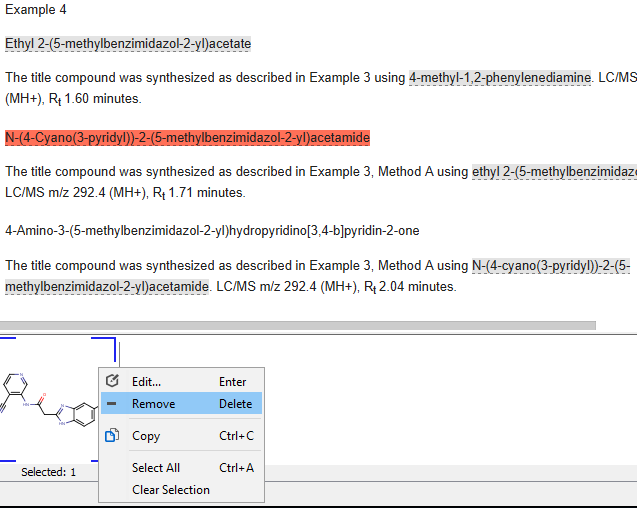
Remove annotations
Any unwanted annotation can be removed by selecting the problematic structure in the document view, and right-clicking on it in the selection view and choosing the Remove option.
Share and Export functions
ChemCurator offers multiple options for project sharing and exporting the annotated data in various formats.
Share projects with ChemCurator integration server
ChemCurator Integration Server is the standard way to share your project with your colleagues and store them in a central database. For details about server installation, please check the Integration Server Administrator Guide. Additionally you need to configure the server connection details in the ChemCurator desktop application following the corresponding section of the Installation Guide.
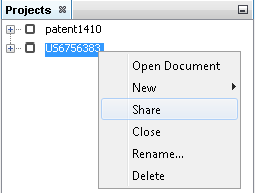
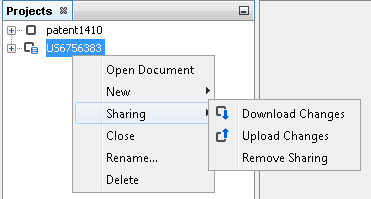
After successful sharing, a new indicator icon appears next to the project, and you are able to upload your modifications or download the newer version of the project.
Export from Compounds and Markush view
Structure export function ![]() is available in compounds and Markush view. The structure and related information from the view can be exported in various file formats.
is available in compounds and Markush view. The structure and related information from the view can be exported in various file formats.
Export project to ZIP file and import from Zip file
A project can be exported to a ZIP file by File>Export Project to ZIP... In this way, the project can easily be shared by e-mail or any file sharing method.
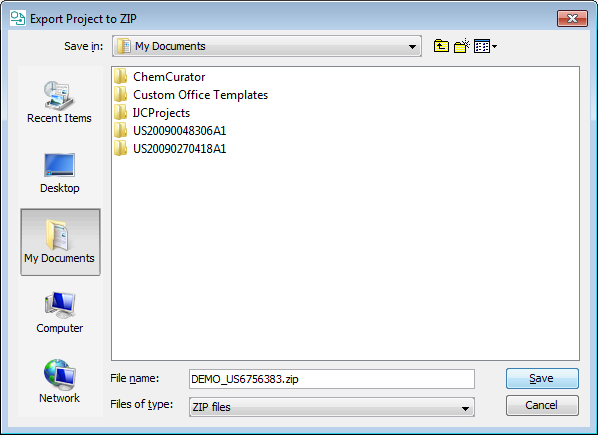
The zipped project can be imported in a similar way using File>Import Project from ZIP... function.
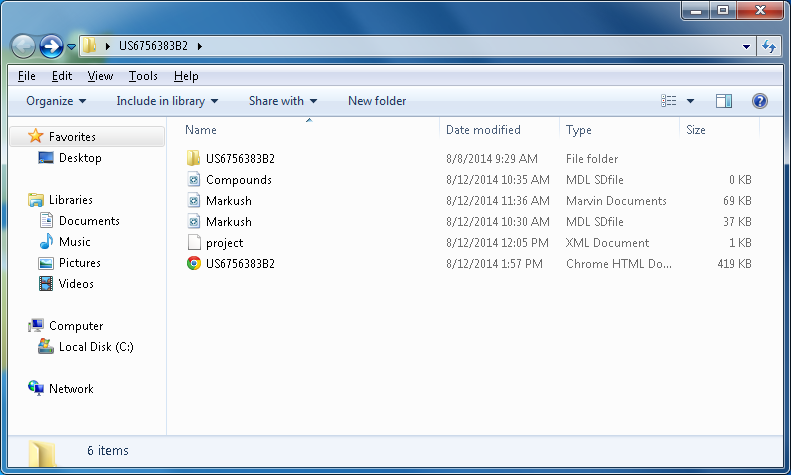
Using the project folder directly
All projects are available in project directories. The default location of the projects is the C:\Users\<user name>\Documents\ChemCurator directory. The name of the project directory always equals to the project name. Every project contains a project file (an XML with some meta data), the annotated document in an HTML file, and (optionally) the associated resources and the extracted chemical information stored in SDF (compound lists) and MRV (Markush structures) formats.