Connect Scripting via Jupyter Notebook¶
- This brief tutorial is created for the demo project. If you're using a different schema, you will need to modify the values accordingly.
- Replace
<YOUR_API_KEY>in the first code snippet with your own API key.
Connecting to Connect session¶
Display all view names¶
Display all views associated to a project.
1 2 3 4 5 6 7 8 9 10 | |
Connect to a View¶
Open view session¶
View session is an object which enables the user to query data and get other information related to the view.
First we connect to the pubchem view session.
Next, let's connect to the wombat view session
Perform query and get data¶
Querying data is based on the jchem query language. Connect API uses the QueryTerm object to perform analogous queries.
Get data tree associated to the pubchem view
Get id field and molweight fields
Query all items where ID field (CdId) has larger value than 1003.
[JUST INPUT TO THE QUERY] Print ID of an ID field.
1 | |
Now retrieve all data which satisfies the query and display first 10 results
1 2 3 4 5 6 7 8 9 10 | |
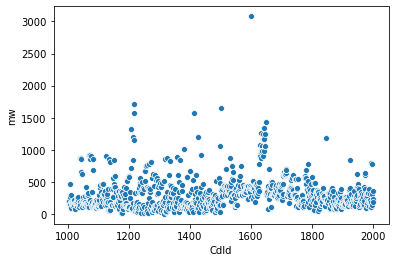
Make a scatter plot of the data. Here I use seaborn library for that purpose
Analyze data¶
1 | |
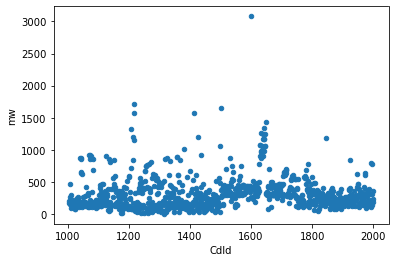
Alternatively this can be done simply by using pandas (both plotting in pandas and seaborn are using an underscoring matplotlib library. However the implementation and settings are different, therefore different default aesthetics.)
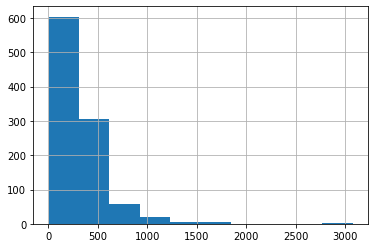
if I want to look at the distribution of molecular weights, then this can be achieved again by using pandas library's methods.
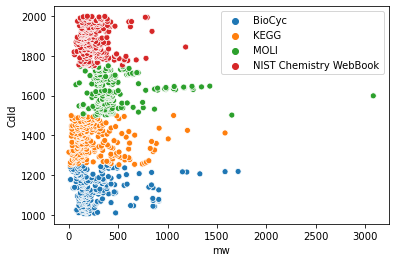
Let's look at the original databases and their relations to molecular weigths. First, we check how many molecules belong to what database.
1 2 3 4 5 | |
Then we display this using a scatterplot
1 | |
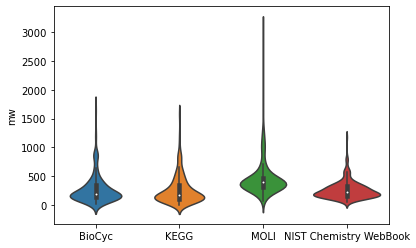
We can see here, that molecules from all four constituing databases have similar molecular weights. However, to really check that, we need different vizualization. Violin plot might come in handy
1 | |
Now, we could say that the mass composition of molecules from the databases is indeed very similar with one possible exception being the MOLI database.
Perform query in wombat¶
Let's begin by displaying all fields.
1 | |
And why not also retrieve all fields?
output
Now, I would like to take all the data and turn it into pandas dataframe object. (This may take a little while).
Let's keep using 'CdId' as an index column instead of creating new one. Also, let's display first five rows:
| Num assay vals | Formula | ID | MOL.REF | VALUE.MIN | BIND.NONSPECIFIC | EST.LOGKOW | BIND.ENDOGENLIGAND | SMDL.ID | SMDL.IDX | ... | SWISSP.SPECIES | REC.TYPE | TARGET.NAME | BIO.EFFECT | Assay values | Rings | MOL.NAME | Mol Weight | logP / MW | REC.FAMILY | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CdId | |||||||||||||||||||||
| 1 | 3 | C18H19ClN2O2 | 1 | Bioorg. Med. Chem. Lett. 12(2)-2002 243-248 | 8.72 | None | 4.98 | None | 470 | SMDL-00000470 | ... | human | None | 5-HT2A | antagonist | IC50 [5-HT2A] 8.7200\nIC50 [5-HT2C] 8.4200\nIC... | 4 | 1 | 330.810 | 66.427711 | GPCR |
| 1 | 3 | C18H19ClN2O2 | 2 | Bioorg. Med. Chem. Lett. 12(2)-2002 243-248 | 8.42 | None | 4.98 | None | 470 | SMDL-00000470 | ... | human | None | 5-HT2C | antagonist | IC50 [5-HT2A] 8.7200\nIC50 [5-HT2C] 8.4200\nIC... | 4 | 1 | 330.810 | 66.427711 | GPCR |
| 1 | 3 | C18H19ClN2O2 | 3 | Bioorg. Med. Chem. Lett. 12(2)-2002 243-248 | 6.99 | None | 4.98 | None | 470 | SMDL-00000470 | ... | human | None | H1 | antagonist | IC50 [5-HT2A] 8.7200\nIC50 [5-HT2C] 8.4200\nIC... | 4 | 1 | 330.810 | 66.427711 | GPCR |
| 2 | 3 | C18H20N2 | 1 | Bioorg. Med. Chem. Lett. 12(2)-2002 243-248 | 7.98 | None | 3.35 | None | 471 | SMDL-00000471 | ... | human | None | 5-HT2A | antagonist | IC50 [5-HT2A] 7.9800\nIC50 [5-HT2C] 8.1300\nIC... | 4 | 3 | 264.372 | 78.917017 | GPCR |
| 2 | 3 | C18H20N2 | 2 | Bioorg. Med. Chem. Lett. 12(2)-2002 243-248 | 8.13 | None | 3.35 | None | 471 | SMDL-00000471 | ... | human | None | 5-HT2C | antagonist | IC50 [5-HT2A] 7.9800\nIC50 [5-HT2C] 8.1300\nIC... | 4 | 3 | 264.372 | 78.917017 | GPCR |
5 rows × 44 columns