Actions¶
Convert to Structure by ID from Database¶
With these options, you can specify the database and conversion settings for the following actions:
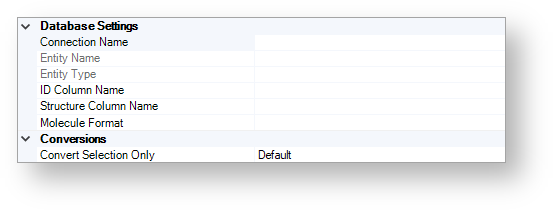
The figure below shows the available database and conversion settings.
Database Setting¶
By using these options, you can specify the database, table, and column from which you would like to import data.
Connection Name¶
All available databases are listed in the Select Entity dialog.
Entity Name¶
This field is automatically filled when a connection selected.
Entity Type¶
This field is automatically filled when a connection selected.
ID Column Name¶
Use the combo box to specify the name of the ID column.
Structure Column Name¶
Use the combo box to specify the name of the structure column.
Molecule Format¶
If you know the format of the structures to be imported, you can specify it by using the drop-down list. This speeds up the import process.
By default, the format is not specified.
Conversions¶
Convert Selection Only¶
Use this option to specify whether the Selection only box is automatically checked or not.
The possible values for this option are as follows:
TRUE
FALSE
DEFAULT
The default value is DEFAULT, which is equal to FALSE.
Convert to Structure by ID from Compound Registration¶
With these options, you can specify the connection and parameter settings for the Resolve CompReg IDs into Structures action.
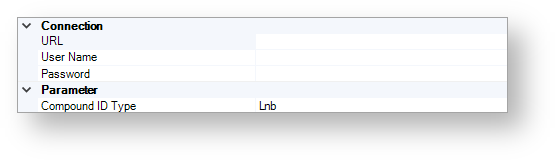
The figure below shows the available connection and parameter settings.
Connection¶
URL¶
Specify the URL of the CompReg database.
User Name¶
Specify the user name which can be used for login.
Password¶
Specify the password that is associated with the user name.
Parameter¶
Compound ID Type¶
Use this option to specify the compound ID type. The specified type must match the ID types on your sheet.
The following types are available:
Lnb
LN
CN
PCN
LotId
StructId
Convert HELM to Structure or Structure to HELM by Biomolecule Toolkit¶
With these options, you can specify the database and conversion settings for the following actions:
- ConvertHelmToStructureAction
- ConvertStructureToHelmAction
in Conversions
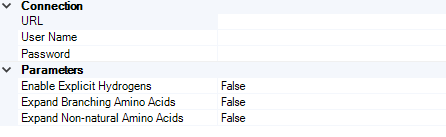
The figure below shows the available database and conversion settings.
Connection¶
URL¶
Specify the URL of the HELM to Structure - Structure to HELM service, http and secure https can be handled as well.
User Name¶
Specify the user name which can be used for login.
Password¶
Specify the password that is associated with the user name.
Parameters¶
Compound ID Type¶
Use this option to specify the default output display of peptide structures after the conversion.
The following types are available:
Enable Explicit Hydrogens
Expand Branching Amino Acids
Expand Non-natural Amino Acids
The default type is False to all 3 options. The detailed description of these parameters are in the documentation of the Biomolecule Toolkit.
Convert from Structure¶
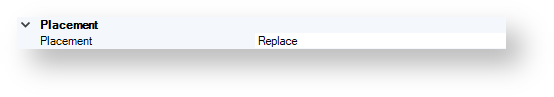
The figure below shows the available convert from structure options.
Placement¶
Placement¶
By using this option, you can specify whether the results of the conversion process replace the structures or not.
The available values are as follows:
Replace
InsertRight
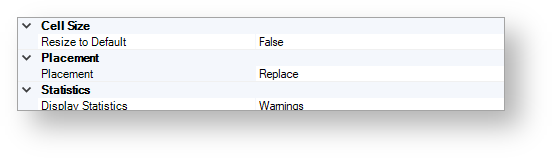
Convert to Structure¶
The figure below shows the available convert to structure options.
Cell Size¶
Resize to Default¶
By using this option, you can specify whether cells are resized to default before the process or not.
The available values for this option are as follows:
TRUE
FALSE
Placement¶
Placement¶
By using this option, you can specify whether the results of the conversion process replace the texts or not.
The available values are as follows:
Replace
InsertRight
Statistics¶
Display Statistics¶
By using this option, you can specify the statistics display behavior.
The following values are available.
On
Off
Warnings
The default value is Warnings. In this case, only issues are displayed, but no conversion report is generated.