Converting implicit Hydrogens to explicit
The following methods are available in chemaxon.calculations.Hydrogenizeclass to convert implicit hydrogen atoms to explicit ones:
Hydrogenize.convertImplicitHToExplicit(MoleculeGraph molecule, MolAtom[] atoms, int f)
Hydrogenize.convertImplicitHToExplicit(MoleculeGraph molecule)
In the first method the coordinate refinement to avoid atom collisions can be skipped using the OMIT_POSTCLEAN option.
You can convert implicit Hydrogens to explicit ones without additional cleaning:
| //import a simple chain
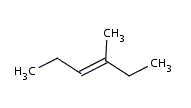
Molecule mol = MolImporter.importMol("methylhexene.mol");
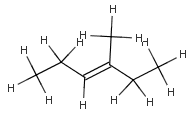
Hydrogenize.convertImplicitHToExplicit(mol, null, MoleculeGraph.OMIT_POSTCLEAN);
|
 |
 |
 |
| original methylhexene molecule |
with OMIT_POSTCLEAN option |
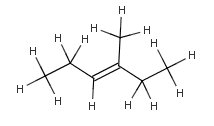
without OMIT_POSTCLEAN option |
Working example to add explicit H to chiral centers only
| import java.io.IOException;
import chemaxon.struc.Molecule;
import chemaxon.struc.MolAtom;
import chemaxon.formats.MolImporter;
import chemaxon.struc.StereoConstants;
public class ExplicitHToChiralCenter {
/**
* Example to add Explicit H to Chiral centers only.
* @param args command line arguments
* @throws java.io.IOException
*
* @version 5.1 04/24/2008
* @since Marvin 5.1
* @author Andras Volford
*/
// ODD and EVEN parity values
static int ODD = StereoConstants.PARITY_ODD;
static int EVEN = StereoConstants.PARITY_EVEN;
public static void main(String[] args) throws IOException {
if (args.length < 1) {
System.err.println("Usage: java ExplicitHToChiralCenter
filename");
System.exit(0);
}
// create importer for the file argument
String s = (String) args[0];
MolImporter molimp = new MolImporter(s);
// store the imported molecules in m
Molecule m = new Molecule();
while (molimp.read(m)) { // read molecules from the file
int ac = m.getAtomCount();
// Atoms with odd or even parity
MolAtom[] t = new MolAtom[ac];
int n = 0;
for (int i = 0; i < ac; i++){
int p = m.getParity(i);
boolean add = p == ODD || p == EVEN;
// if the atom has ODD or EVEN parity
if (add) {
t[n++] = m.getAtom(i);
}
}
// reduce atom array
MolAtom[] a = new MolAtom[n];
System.arraycopy(t, 0, a, 0, n);
// add explicit H
Hydrogenize.convertImplicitHToExplicit(m, a, 0);
if (m.getDim() != 2)
m.clean(2, null);
// write the result
System.out.print(m.toFormat("sdf"));
}
}
}
|


