DB Web Services¶
- Microservices system mode
- Standalone web application mode
- Download
- System requirements
- Installation
- Licenses
- Logging
- Configuration
- Timeout handling
- High Availability (HA)
- Running the server
- API Documentation
- Demo site
- Usage
- Data recovery
- Import/Export tables using S3 bucket
- AWS Fargate setup
- Performance measurement and recommended system setup
DB Web Services had a major update in version 25.3.0. If you are using en earlier version, read this documentation instead, or see our upgrade guide.
DB Web Services provides the functionality to store and search chemical structures in a persistent database, currently in H2 and PostgreSQL databases. It has endpoints for:
- creating/deleting tables,
- inserting/deleting/modifying structures and data in the tables,
- executing duplicate, substructure, full fragment and similarity searches.
This documentation describes installation, administration and usage of DB Web Services. As all JChem Microservices modules, it is available in two modes:
- As part of a microservices system
- As a standalone web application
Microservices system mode¶
In microservices system mode, the DB Web Services runs together with the Config, Discovery and Gateway services. These three services are mandatory, and optionally other services can also be part of the system. All configuration must be done in the Config service.
The default configuration applies to the microservices system mode.
The web application runs on host <server-host> and listens on port <gateway-server-port>.
Standalone web application mode¶
In standalone web application mode, the DB Web Services runs alone, without the Config, Discovery and Gateway services (however, the installer installs them as well).
The default configuration must be changed according to the standalone web application mode. Set the following in the application.properties file
eureka.client.enabled=falsespring.cloud.config.enabled=false- If you are using a version earlier than 25.3.0 remove the line that starts with
spring.config.import=
All configuration must be done in the DB module.
The web application runs on host <server-host> and listens on port <server-port>.
Download¶
See here.
System requirements¶
See here
Installation¶
See here.
Module is installed into folder: jws/jws-db
Licenses¶
See here.
Logging¶
See here.
Configuration¶
Default configuration:
| application.properties | description |
|---|---|
| server.port=8062 | |
| logging.file.name=../logs/jws-db.log | |
| spring.config.import=optional:configserver:${CONFIG_SERVER_URI:http://localhost:8888} | Config service connection configuration with retry mechanism options. |
| eureka.client.enabled=true | set eureka.client.enabled=false to switch to standalone DB Web Services application mode |
| initOnStart=${DB_INIT_ON_START:AUTO} | initOnStart can be:INIT: the existing database is deleted, and a new empty one is createdAUTO: existing database is started, in case of non-existing database a new empty one is created |
| updateMode=${DB_UPDATE_MODE:EXIT} | updateMode gets in action only if the version number has changed updateMode can be EXIT: exit if version mode has changedDROP: drop old data if version mode has changedREINDEX: keep old data and reindex them in order to work with new versionFORCE_REINDEX: keep old data and reindex them regardless of version change |
| chemaxon.microservices.db.cache.enabled=true | Use in-memory cache to improve performance of search functionalities. Resource requirement and configuration details are available in JChem Engines cache and memory calculator page |
| chemaxon.microservices.db.cache.similarity.enabled=true | Enable in-memory caching to improve performance of tautomer similarity search. Resource requirement and configuration details are available in JChem Engines cache and memory calculator page |
| chemaxon.microservices.db.datasource.h2.dir=${CXN_STRUCTURE_DATA_DIR:./data/chemical-data/store} | Stores database files - used only in case of H2 database. |
| chemaxon.microservices.db.datasource.dialect=${CXN_DB_DIALECT:H2} chemaxon.microservices.db.datasource.url= ${CXN_DB_JDBC_URL:jdbc:h2:nio:${chemaxon.microservices.db.datasource.h2.dir}/db;COMPRESS=true;MAX_COMPACT_TIME=10000;DEFRAG_ALWAYS=TRUE} chemaxon.microservices.db.datasource.username=${CXN_DB_JDBC_USER:user} chemaxon.microservices.db.datasource.password=${CXN_DB_JDBC_PASSWORD:password} |
Database type and connection configurations |
| chemaxon.microservices.db.import-export.dir=${CXN_DB_IMPORT_EXPORT_DIR:data/export} chemaxon.microservices.db.import-export.import-batch-size=${CXN_DB_IMPORT_EXPORT_BATCH_SIZE:5000} |
Folder name where DB can be exported/imported and importing batch size. |
| chemaxon.microservices.db.types[0].type-name = sample chemaxon.microservices.db.types[0].type-id = 1 chemaxon.microservices.db.types[0].tautomer-handling-mode = OFF chemaxon.microservices.db.types[0].stereo-assumption=ABSOLUTE chemaxon.microservices.db.types[0].standardizer-config = aromatize chemaxon.microservices.db.types[0].fp-length-in-bits = 512 chemaxon.microservices.db.types[0].fp-edges = 6 chemaxon.microservices.db.types[0].fp-ones = 2 chemaxon.microservices.db.types[0].canonical-tautomer-heavy-atom-limit = 100 chemaxon.microservices.db.types[1].type-name = taumol chemaxon.microservices.db.types[1].type-id = 2 chemaxon.microservices.db.types[1].tautomer-handling-mode = GENERIC chemaxon.microservices.db.types[1].stereo-assumption=ABSOLUTE chemaxon.microservices.db.types[1].standardizer-config = aromatize chemaxon.microservices.db.types[1].fp-length-in-bits = 512 chemaxon.microservices.db.types[1].fp-edges = 6 chemaxon.microservices.db.types[1].fp-ones = 2 chemaxon.microservices.db.types[1].canonical-tautomer-heavy-atom-limit = 100 |
Molecule types define the interpretation mode of the chemical structures. Important: Indexing of the types array must always be sequential from 0: 0, 1, 2, ... typeName must be uniquetypeID must be unique integer tautomerHandlingMode can be OFF GENERIC NORMAL_CANONICAL_GENERIC_HYBRIDNORMAL_CANONICAL_NORMAL_GENERIC_HYBRIDstereoAssumption can be ABSOLUTE or RELATIVEstandardizerConfig can be made of action strings standardizerConfig=aromatize:b..removeExplicitHThe standardizer configuration can be specified also as chemaxon.microservices.db.types[n].standardizerFile but strictly use only one of them, standardizerConfig OR standardizerFile. If neither standardizerConfig, nor standardizerFile is specified (or both are empty), then we use the default standardizer config, instead, which is “aromatize” fpLengthInBits must be a positive multiple of 64. The default is 512. Setting a higher value increases indexing time, but may give better results in similarity search. canonicalTautomerHeavyAtomLimit is available limiting the size of molecules the tautomer form of which is taken into account in duplicate and fullfragment search in the case of NORMAL_CANONICAL_GENERIC_HYBRID tautomerHandlingMode.New molecule type can be introduced for existing database. Any modification or delete changes take effect only if initOnStart is set to INIT, and the application is re-started, but take care, the existing database will be deleted. |
Search logging
Debug level of search logging can be set in the files configuring the JVM options:
jws-db-service.vmoptions
run-jws-db.vmoptions
by adding line
-Djchem.debug=true
or setting
in application.properties file.
Timeout handling¶
Timeout limit can be provided as an optional parameter in different structure insertion and search requests but default timeout limit also can be configured with application properties.
Structure insertion¶
Timeout handling attributes can be used in following endpoints
/rest-v1/db/additional/upload/rest-v1/db/additional/{tableName}/batchInsert
| attribute name | attribute type | attribute place | description |
|---|---|---|---|
| timeoutInMilliseconds | integer | request parameter | Optional request parameter. Timeout limit in milliseconds. |
| offset | integer | request parameter | Optional request parameter. Structure insertion starts from this structure of file/request. |
| stoppedByTimeout | boolean | response attribute | It is true when insert process is interrupted due timeout. |
| processed | integer | response attribute | Number of processed elements from the request. The structure loading is done sequentially. If process is interrupted maybe not all structures are inserted. offset can be calculated based on it for the next insert request |
When timeoutInMilliseconds is not provided, the default timeout limit is determined by the spring.cloud.gateway.server.webflux.httpclient.response-timeout application property.
Structure search¶
Timout limit can be provided in all type of duplicate, full fragment, similarity and substructure table searches with timeoutInMilliseconds request parameter.
When timeoutInMilliseconds is not provided, the default timeout limit is determined by the spring.cloud.gateway.server.webflux.httpclient.response-timeout application property.
Configuration¶
Timeout limit can be controlled by following application properties.
| application.properties | description |
|---|---|
| spring.cloud.gateway.server.webflux.httpclient.response-timeout=25000 | The default timeout limit for structure insertion and search, in millisecond, when related parameter is not provided. |
| chemaxon.microservices.db.timeout.stop-earlier=1000 | DB Web Services interrupt insert or search requests this number of milliseconds before the timeout limit to ensure a response can be provided to the gateway in time. |
High Availability (HA)¶
HA and load balancing is provided for DB Web Services. Running more instances of the db service ensures HA and load balancing.
Requirements¶
HA mode needs PostgreSQL database. It is not supported with H2. As DB Web Services instances do not use distributed cache no additional configuration needed.
Load balanced example¶
Here you find a load balanced example application on GitHub.
Running the server¶
Prerequisites in case of microservices system mode:
- Config service is running
- Discovery service is running
- Gateway service is running
Run the service in command line in folder jws/jws-db/ :
jws-db-service.exe --install
jws-db-service.exe --start (on Windows in administrator's terminal)
jws-db-service start (on Linux)
or
run-jws-db.exe (on Windows)
run-jws-db (on Linux)
API Documentation¶
Find and try out the API on the Swagger UI.
| Mode | URL of Swagger UI | default URL of Swagger UI |
|---|---|---|
| microservices system | <serverhost>:<gateway-port>/jwsdb/API/ | localhost:8080/jwsdb/API/ |
| standalone web application mode | <serverhost>:<server-port>/API/ | localhost:8062/API/ |
Demo site¶
For detailed description check out the JWS DB demo site:
https://jchem-microservices.chemaxon.com/jwsdb/api/index.html
Usage¶
The guidelines, examples on the Demo site or on the Swagger UI API documentation of your installed module display the methods and syntax implemented for reaching the essential chemical searching functionalities of JChem Base.
Molecule type information¶
DB Web Services provides method for getting the available molecule types.
Every table has a Molecule type: this is a descriptor that is used by the search engine. It contains information about how structures are handled during search. The application has two very simple built in types called: sample (search with aromatization) and taumol (tautomer search). See the type definitions in the application.properties file.
Store and search molecules and non-chemical data¶
Table operations
Structure Insert/Delete methods
Duplicate search methods
Substructure search methods
Similarity search methods
Search on additional data¶
DB Web Services provide additional data filtering option in POST request case on below endpoints.
Additional data filtering is executed after chemical filter so search performance is better if query molecule is well-defined and narrow the result set.
/rest-v1/db/additional/{tableName}/fullfragment/rest-v1/db/additional/{tableName}/similarity/rest-v1/db/additional/{tableName}/substructure
Possible filtering options in additionalDataCondition attribute
Text¶
Text additional data can be filtered with following operators.
exact- Filter text is exactly the same as additional data valuecontains- Additional data contains the provided filter textnotExact- Filtered additional data type is defined on molecule and has different value than filter text
Number¶
Number can be filtered with following operators: >, >=, <, <=, =, !=
Complex¶
Multiple filters can be combined with and and or logical operators
Data recovery¶
The data is stored in PostgreSQL database server or in H2 database case in the file system under the folder which is configred in chemaxon.microservices.db.datasource.h2.dir application property. The content of that folder must be stored as backup. Furthermore, the application.properties file(s) also should be saved.
Import/export tables using S3 bucket¶
It is possible to use S3 buckets to import/export database tables. For this we use Spring Could AWS.
Our default settings are different than Spring's defaults so users who are not using this feature can run their services without AWS issues.
The current constant is the following:
| application.properties | description |
|---|---|
| spring.cloud.aws.region.static=eu-central-1 | A static value for region used by auto-configured AWS clients |
Credential configuration details are available in official Spring Cloud AWS documentation.
You can setup S3 import/export the following settings:
AWS Fargate setup¶
When running DB Web Services on AWS Fargate, it is recommended to use a persistent data storage (e.g. Amazon RDS) and if upgrading in REINDEX mode, the export/import strategy should be set to S3:
Because in REINDEX mode the service reimports the tables at startup, the timeout and startPeriod of the AWS Fargate service should be set accordingly so it would give enough time for the reimport. (AWS API Reference - ECS HealthCheck)
The above import/export settings for S3 should also be set if using /rest-v1/db/additional/{tableName}/importFromFile/{fileName} and /rest-v1/db/additional/{tableName}/exportToFile endpoints on AWS Fargate.
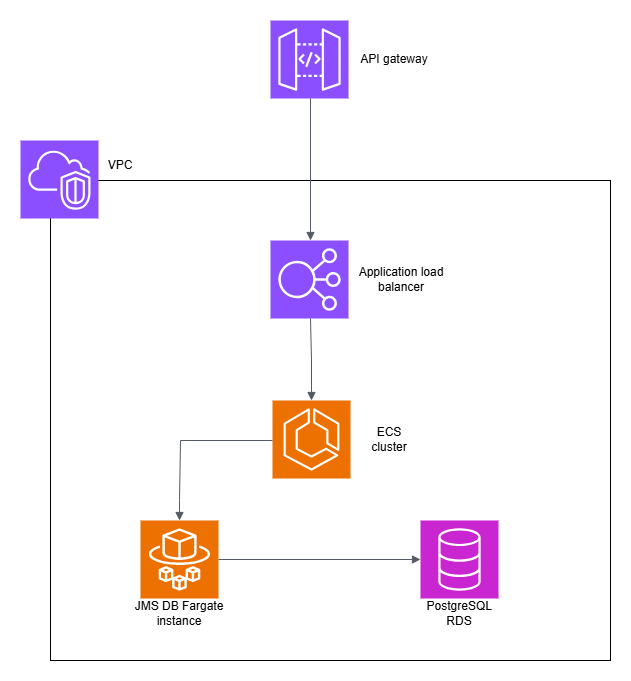
Setting up backend suitable for AWS Fargate¶
DB Web Services use a backend where index data is stored through a JDBC connection. As default this JDBC connection is a file based H2 connection. On AWS Fargate systems persisting data on the file system of the AWS Fargate container is not advised as after deleting the container data gets lost. Therefore, we suggest using a database outside the AWS Fargate container.
An example for setting a PostgreSQL Amazon RDS instance:
JSON Converter¶
Converts different molecular string representations into JSON format and molecular JSON representations to string.
Performance measurement and recommended system setup¶
The search performance of DB Web Services was evaluated using datasets of various sizes in an AWS environment.
The objective was to identify a cost-optimized system configuration that maintains stable performance under high workloads.
Environment¶
All measurements were conducted in Amazon Web Services (AWS).
- DB Web Services ran in standalone mode using the ECS Fargate service.
- AWS RDS was used to host PostgreSQL databases 17.6 version.
- The DB Web Services instance had its cache preloaded prior to measurement.
Measurement process¶
The following DB Web Services endpoints were tested using the POST method:
- /rest-v1/db/additional/{tableName}/duplicate
- /rest-v1/db/additional/{tableName}/similarity
- /rest-v1/db/additional/{tableName}/substructure
For each search type, 30–50 representative query molecules were selected to cover common use cases.
Substructure searches were tested with different query sets, categorized by hit frequency (very frequent, frequent, rare) and special functionality (stereochemistry).
Search results were limited to 10 or 100 hits.
Performance was measured in two scenarios:
- Sequential — queries sent one by one.
- Parallel — queries sent concurrently from 5 clients.
Databases were created from randomly selected subsets of the Enamine dataset with the following structure counts:
- 1.000.000
- 10.000.000
- 100.000.000
- 500.000.000
- 1.000.000.000
All tests were performed using the default "sample" molecule type, so tautomers were not considered.
The measurement process was executed using the K6 framework.
Measurement result¶
Suggested environment configuration according measurement result.
| structure count | service vCPU | service RAM | service Xmx | DB type | DB vCPU | DB RAM |
|---|---|---|---|---|---|---|
| 1 million | 1 | 4 GB | 2580m | db.t3.micro | 2 | 1 GB |
| 10 million | 1 | 4 GB | 3292m | db.t3.micro | 2 | 1 GB |
| 100 million | 2 | 12 GB | 10420m | db.t3.small | 2 | 2 GB |
| 500 million | 8 | 44 GB | 44100m | db.m7g.large | 2 | 8 GB |
| 1 billion | 16 | 96 GB | 88000m | db.r5.xlarge | 4 | 32 GB |
Measurement was executed with 25.3.1 version.
Result details are available in the file jms_standardized_measurement_25_3_1.xlsx.
This file includes measurement result sheets — performance metrics by dataset size.
Results may vary by 5–10%, especially under parallel load conditions.
These measurements cover the typical search scenarios. However, there can be some specific cases which are not handled with the same efficiency by the search engine and measurement results can be significantly different.
Structure upload performance¶
Databases were created from molecule files, each containing 1 million structures.
Upload performance primarily depends on the number of available vCPUs and whether duplicate filtering is enabled.
The Upload sheet in jms_standardized_measurement_25_3_1.xlsx contains detailed upload performance metrics.