MarvinView in JTable Example¶
Since chemical structures are frequently stored in databases, the most common usage of molecule visualization is perhaps in a tabular format that allows browsing the database, showing search results, selections of structures against particular criteria.
In typical Java applications, the javax.swing.JTable is used to display tables of data. This form of visualization form will be referred to as tables. This section demonstrates how can molecules be rendered with Marvin inside existing tables.
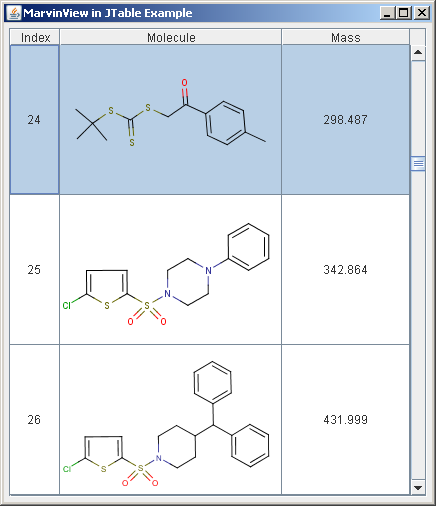
The following picture shows a simple table with a molecule column.
The characteristic features of JTable to bear in mind are as follows:
- JTable does not contain or cache data, it is simply a view of your data.
- Molecule tables can contain a large number of structures, thus speed and the amount of required disk space is critical.
- JTable optionally enables the editing of the structure data.
It is important to understand the concepts of JTable about Editors and Renderers. Let us quote the relevant part of the Java Tutorials:
You might expect each cell in a table to be a component. However, for performance reasons, Swing tables are implemented differently.
Instead, a single cell renderer is generally used to draw all of the cells that contain the same type of data. You can think of the renderer as a configurable ink stamp that the table uses to stamp appropriately formatted data onto each cell. When the user starts to edit a cell's data, a cell editor takes over the cell, controlling the cell's editing behavior.
To choose the renderer that displays the cells in a column, a table first determines whether you specified a renderer for that particular column. If you did not, then the table invokes the table model's getColumnClass method, which gets the data type of the column's cells. Next, the table compares the column's data type with a list of data types for which cell renderers are registered. This list is initialized by the table, but you can add to it or change it.
Chemaxon recommends the following points to consider:
- There is only one single cell renderer that is required to display molecules in tables, regardless of the number of structures being available.
- There are two major approaches provided to create a cell renderer with Marvin: using MViewPane or simply painting the molecules with MolPrinter.
- A cell editor is required only in case the structure itself is allowed to be modified by the user. Otherwise, it is not needed to use a complex component for rendering.
Creating Cell Renderer with MViewPane¶
To use the MViewPane component as the cell renderer, it should implement the getTableCellRendererComponent method of the TableCellRenderer interface. The implementation of this method sets up the rendering component to display the passed-in molecule, and then returns the component.
This renderer class is accessible from the public API.
Creating Cell Renderer with MolPrinter¶
In many cases, it is sufficient to have only the static image of the molecule inside cells, that is, no need to rotate, change rendering mode, or modify the structure itself. The renderer can be more compact in this case.
This renderer extends the JPanel class overriding its paintComponent method. The getTableCellRenderer method sets the molecule to be shown.
There are several ways to specify a cell renderer. In this example, we use type-specific renderers using the setDefaultRenderer method of JTable.
for setting MViewPane renderer, while
for setting MolPrinter renderer.
Creating Cell Editor with MViewPane¶
Using MViewPane as a cell editor provides two major advantages.
- It offers many functionalities that modify the visualization but not the structure itself, for example, rotation, zoom, display and color schemas, or the dimension in which the structure is shown.
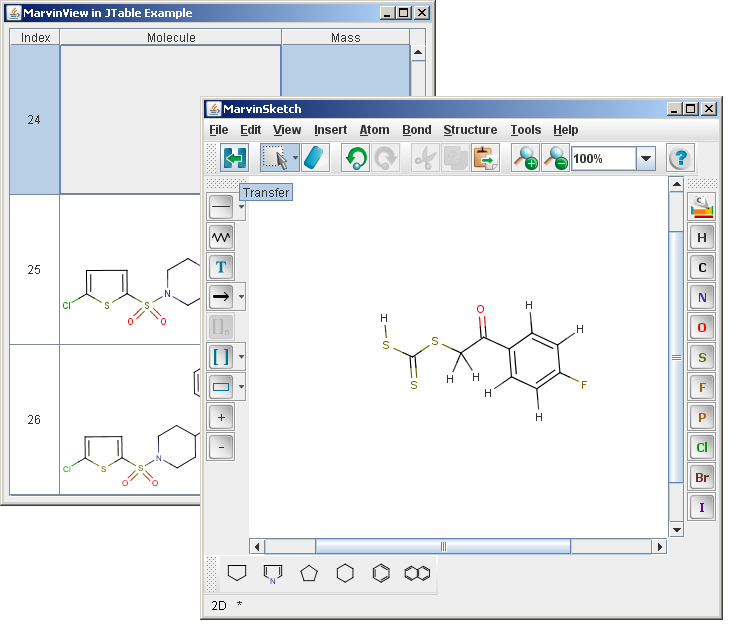
- It can also be used as an indirect cell editor. Double-clicking on the MViewPane cell editor brings up MarvinSketch, in which the structure can be modified. After MarvinSketch is closed, the molecule is updated.
The image below shows indirect editing in use:
The code below shows the major parts of the simple cell editor.
This renderer class is accessible from the public API.
In case the molecular structures can be modified by a cell editor, we can ensure that the modification affects all other data being in a connection, like computed molecular data.
In this example, the third column contains such computed molecular data, the mass.
When the editing of the molecule is finished and fireEditingStopped is called, the setValueAt(Object value, int row, int col) method of the table model gets called. In this example, we have set the molecule masses inside this method, so that when the molecule is initialized and every time it is modified, its mass will be instantly updated.
The code excerpts shown above were taken from the full sample code of
ViewJTable.java,MolRenderer.java,MViewRenderer.java,MViewEditor.java.