Markush structure specific search options
In this document the search options concerning Markush structures are summarized and their usage is shown in different search interfaces.
Homology narrow translation
This option relates to query-side homology handling. The homology narrow translation option can switch off special handling of homology groups, hence they can match the homology group of the same type only (e.g. alkyl on alkyl or chk). The option can have the following values:
-
ALL/a: All atoms can have translation.
-
MARKED/m: Only marked atoms can have translation. (Not implemented yet.)
-
NONE/n: No atom can have translation.
Limitations:
-
If homology narrow translation is allowed than query side homology against Markush targets is limited to those cases when Markush targets do not contain any other Markush features than the same homology group as the guery structure does.
-
Properties of homology groups (e.g. LO, MID, HI on
alkyl) are ignored when both the query and the target structure contains the homology group. -
Rgroups with more than two attachment points are not supported on the query side.
More information about translation options can be found in theMarkush search documentation.
| Query | Target | Homology narrow translation | |
|---|---|---|---|
| None | All | ||
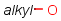 |
 |
 |
 |
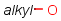 |
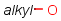 |
 |
 |
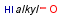 |
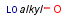 |
 |
 |
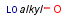 |
 |
 |
 |
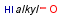 |
 |
 |
 |
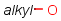 |
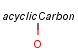 |
 |
 |
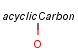 |
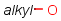 |
 |
 |
MolSearch API
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setHomologyNarrowTranslation( HomologyTranslationOption.NONE /
HomologyTranslationOption.ALL / HomologyTranslationOption.MARKED );
// ...
MolSearch searcher = new MolSearch(); searcher.setSearchOptions(searchOptions);
Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE.JChemSearch API
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setHomologyNarrowTranslation( HomologyTranslationOption.NONE /
HomologyTranslationOption.ALL / HomologyTranslationOption.MARKED );
// ...
JChemSearch searcher = new JChemSearch();
searcher.setSearchOptions(searchOptions);
Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE.JChem Oracle Cartridge
Use the jc_compare operator with homologyNarrowTranslation:n/a/mjcsearch command line tool
Use the following command line parameter:
--homologyNarrowTranslation:n/a/mHomology broad translation
Handling of target side homology groups is controlled by the homology broad translation option. If matching is switched off for a query atom, then this query atom can match homology atom only if it is the same homology atom. If matching is switched on for a query atom it matches homology group representing a larger set of structures. E.g. acyclic carbon atom can match alkyl or carbontree, an Fe atom can match transition metal or metal homology atom. Homology atoms can also match homology atoms covering a larger set of structures: carboalicyclyl can match cyclyl or xx. In this cases the query homology atom is a subset of the target homology atom. Subset rules can be seen here. Possible values for homology translation: The option can have the following vaules:
-
ALL/a: all query atoms can match on broader homology atoms, if the properties of the group are fulfilled.
-
MARKED/m: only the marked query atoms can match on broader homology groups. This option is not yet implemented it works as none.
-
NONE/n: no query atoms can match on broader groups, they can only match if the query atom is a homology atom of the same group.
Default value is NONE. Further properties of translation NONE behavior:
-
homology groups cannot match and cannot be matched by specific atoms or homology groups being a superset or a subset of the given group;
-
homology groups can match pseudo atoms with alias name of the given group (e.g. chk matches alkyl);
-
homology properties are ignored (alkyl,LO matches alkyl,HI).
Read more about homology groups.
| Query | Target | Homology broad translation | |
|---|---|---|---|
| None | All | ||
 |
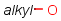 |
 |
 |
 |
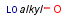 |
 |
 |
 |
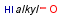 |
 |
 |
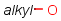 |
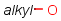 |
 |
 |
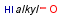 |
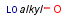 |
 |
 |
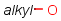 |
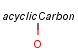 |
 |
 |
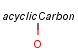 |
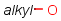 |
 |
 |
MolSearch API
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setHomologyBroadTranslation( HomologyTranslationOption.NONE /
HomologyTranslationOption.ALL / HomologyTranslationOption.MARKED );
// ...
MolSearch searcher = new MolSearch();
searcher.setSearchOptions(searchOptions);
Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE.JChemSearch API
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setHomologyBroadTranslation( HomologyTranslationOption.NONE /
HomologyTranslationOption.ALL / HomologyTranslationOption.MARKED );
// ...
JChemSearch searcher = new JChemSearch();
searcher.setSearchOptions(searchOptions);
Default value is HomologyTranslationOption.MARKED, but it is not implemented yet and works as HomologyTranslationOption.NONE.JChem Oracle Cartridge
Use the jc_compare operator with homologyBroadTranslation:n/a/mjcsearch command line tool
Use the following command line parameter:
--homologyBroadTranslation:n/a/mMarkush search enable/disable
Sets whether targets containing Markush features should be treated as Markush libraries or not. True - if a target containing Markush features should be treated as Markush library. Default value is false.
MolSearch API
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setMarkushEnabled( true / false );
// ...
MolSearch searcher = new MolSearch();
searcher.setSearchOptions(searchOptions);JChemSearch API
Cannot be set. Depends on database table type.JChem Oracle Cartridge
Not applicable.jcsearch command line tool
Use the following command line parameter:
--markush:n/y
Default value is n. Complete homology groups
Sets if only complete structures should match on homology groups. See detailed explanation here. MolSearch API
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setCompleteHG( true / false );
// ...
MolSearch searcher = new MolSearch();
searcher.setSearchOptions(searchOptions);
Default value is true.JChemSearch API
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setCompleteHG( true / false );
// ...
JChemSearch searcher = new JChemSearch();
searcher.setSearchOptions(searchOptions);
Default value is true.JChem Oracle Cartridge
Use the jc_compare operator with completeHG:y/njcsearch command line tool
Use the following command line parameter:
--completeHG:y/nMarkush screening
Specifies whether screening should be used in case of Markush search. Default value is true or y. MolSearch API
Not applicable.JChemSearch API
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setMarkushScreeningEnabled( true / false );
// ...
JChemSearch searcher = new JChemSearch();
searcher.setSearchOptions(searchOptions);
Default value is true.JChem Oracle Cartridge
Not applicable.jcsearch command line tool
Use the following command line parameter:
--markushScreening:y/n Markush fingerprint screening
Specifies whether fingerprint screening should be used in case of Markush search. Default value is true. MolSearch API
Not applicable.JChemSearch API
JChemSearchOptions searchOptions = new JChemSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setMarkushFPScreeningEnabled( true / false );
// ...
JChemSearch searcher = new JChemSearch();
searcher.setSearchOptions(searchOptions);JChem Oracle Cartridge
Not applicable.jcsearch command line tool
Not applicable.Hit-index type
Sets the representation to use for hit indexes in case of Markush search. MolSearch API
MolSearchOptions searchOptions = new MolSearchOptions(SearchConstants.SUBSTRUCTURE);
searchOptions.setHitIndexType( SearchConstants.MARKUSH_HIT_ORIGINAL / SearchConstants.MARKUSH_HIT_INNER );
// ...
MolSearch searcher = new MolSearch();
searcher.setSearchOptions(searchOptions);
Default value is SearchConstants.MARKUSH_HIT_ORIGINAL.JChemSearch API
Not applicable.JChem Oracle Cartridge
Not applicable.jcsearch command line tool
Use the following command line parameter:
--hitIndexType:m/i
m: (default) returns hits for the original Markush diagram;
i: (default) returns hits for the inner compiled representation.